Boiani M., Parpinelli R. S. A GPU-based hybrid jDE algorithm applied to the 3D-AB protein structure prediction. Swarm and Evolutionary Computation (2020): 100711. doi: 10.1016/j.swevo.2020.100711, URL
Boiani M., Dominico G., Parpinelli R. S. (2019) A GPU-Based Hybrid jDE Algorithm Applied to the 2D-AB Protein Structure Prediction Problem. 2019 8th Brazilian Conference on Intelligent Systems (BRACIS), Salvador, Brazil, 2019, pp. 317-322, doi: 10.1109/BRACIS.2019.00063, URL
Boiani M., Dominico G., Parpinelli R. S. (2020) A GPU-Based jDE Algorithm Applied to Continuous Unconstrained Optimization. In: Abraham A., Cherukuri A., Melin P., Gandhi N. (eds) Intelligent Systems Design and Applications. ISDA 2018. Advances in Intelligent Systems and Computing, vol 940. Springer, Cham. doi: 10.1007/978-3-030-16657-1_85, URL
Description: Protein Structure Prediction (PSP) problem is an open problem in bioinformatics and, as the problem scales, complexity and processing time increases. In this way, robust methods and massively parallel architectures are required. This repository provide a GPU-based hybrid algorithm, named cuHjDE, to handle the 3D-AB off-lattice PSP problem. The cuHjDE is composed of the jDE algorithm and the Hooke-Jeeves local search algorithm. An important feature present in the proposed method is the use of a crowding mechanism to avoid premature convergence promoting diversification in the search space.
-
CUDA Toolkit (tested with 9.2)
-
Boost C++ Libraries - Program Options (tested with 1.58.0)
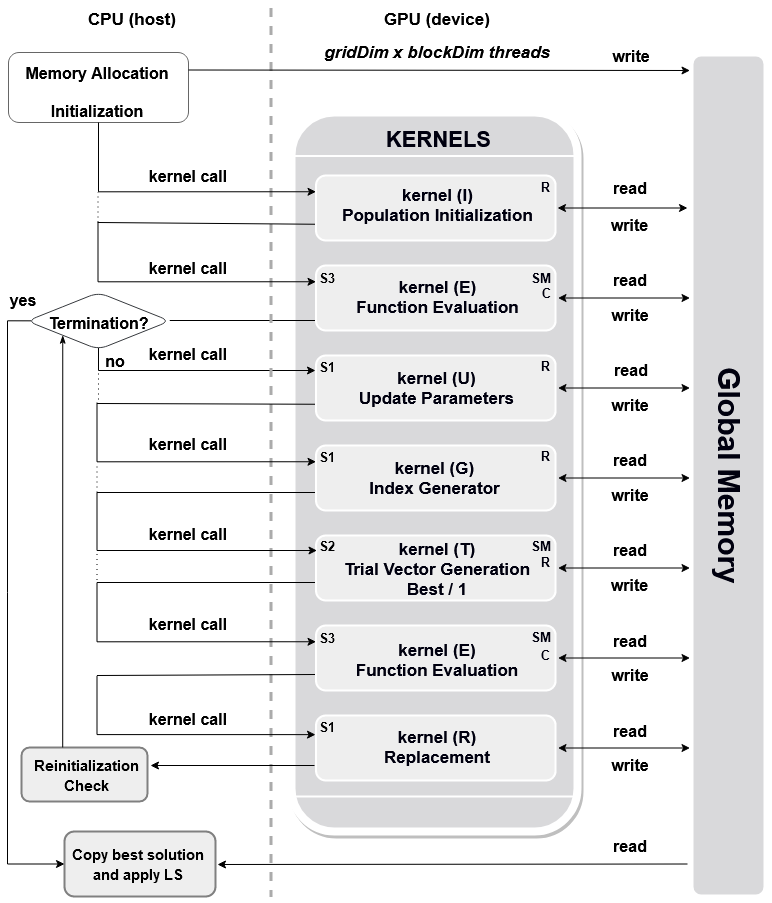
The Figure below depicts a flow chart of the GPU-based implementation. In kernels’ boxes, the labels S1, S2, and S3 represent the kernel structure and letters R,SM, and C denote the use of random number generator, shared memory and constant memory, respectively
$ cd repo
$ make$ "runs, r" - Number of Executions
$ "pop_size, p" - Population Size
$ "dim, d" - Protein Length {13, 21, 34, 38, 55, 64, 98, 120}
$ "max_eval, e" - Number of Function Evaluations
$ "help, h" - Show this help
| PDB ID | PSL | D | AB Sequence |
|---|---|---|---|
| 1BXP | 13 | 21 | ABBBBBBABBBAB |
| 1CB3 | 13 | 21 | BABBBAABBAAAB |
| 1BXL | 16 | 27 | ABAABBAAAAABBABB |
| 1EDP | 17 | 29 | ABABBAABBBAABBABA |
| 2ZNF | 18 | 31 | ABABBAABBABAABBABA |
| 1EDN | 21 | 37 | ABABBAABBBAABBABABAAB |
| 2H3S | 25 | 45 | AABBAABBBBBABBBABAABBBBBB |
| 1ARE | 29 | 53 | BBBAABAABBABABBBAABBBBBBBBBBB |
| 2KGU | 34 | 63 | ABAABBAABABBABAABAABABABABABAAABBB |
| 1TZ4 | 37 | 69 | BABBABBAABBAAABBAABBAABABBBABAABBBBBB |
| 1TZ5 | 37 | 69 | AAABAABAABBABABBAABBBBAABBBABAABBABBB |
| 1AGT | 38 | 71 | AAAABABABABABAABAABBAAABBABAABBBABABAB |
| 1CRN | 46 | 87 | BBAAABAAABBBBBAABAAABABAAAABBBAAAAAAAABAAABBAB |
| 2KAP | 60 | 115 | BBAABBABABABABBABABBBBABAABABAABBBBBBABBBAABAAABBABBABBAAAAB |
| 1HVV | 75 | 145 | BAABBABBBBBBAABABBBABBABBABABAAAAABBBABAABBABBBABBAABBABBAABBBBBAABBBBBABBB |
| 1GK4 | 84 | 163 | ABABAABABBBBABBBABBABBBBAABAABBBBBAABABBBABBABBBAABBABBBBBAABABAAABABAABBBBAABABBBBA |
| 1PCH | 88 | 171 | ABBBAAABBBAAABABAABAAABBABBBBBBABAAABBBBABABBAABAAAAAABBABBABABABABBABBAABAABBBAABBAAABA |
| 2EWH | 98 | 191 | AABABAAAAAAABBBAAAAAABAABAABBAABABAAABBBAAAABABAAABABBAAABAAABAAABAABBAABAAAAABAAABABBBABBAAABAABA |
$ cd repo
$ ./demo <parameter setting> or make run (with default parameters)$ make clean- Empty list for a while
[1] J. Brest, V. Zumer and M. S. Maucec, "Self-Adaptive Differential Evolution Algorithm in Constrained Real-Parameter Optimization," 2006 IEEE International Conference on Evolutionary Computation, Vancouver, BC, 2006, pp. 215-222. doi: 10.1109/CEC.2006.1688311, URL
[2] CUDA is a parallel computing platform and programming model developed by NVIDIA for GPGPU