This is the git repository for the FUNDER project and base for the data paper: xxx et al. (not written yet).
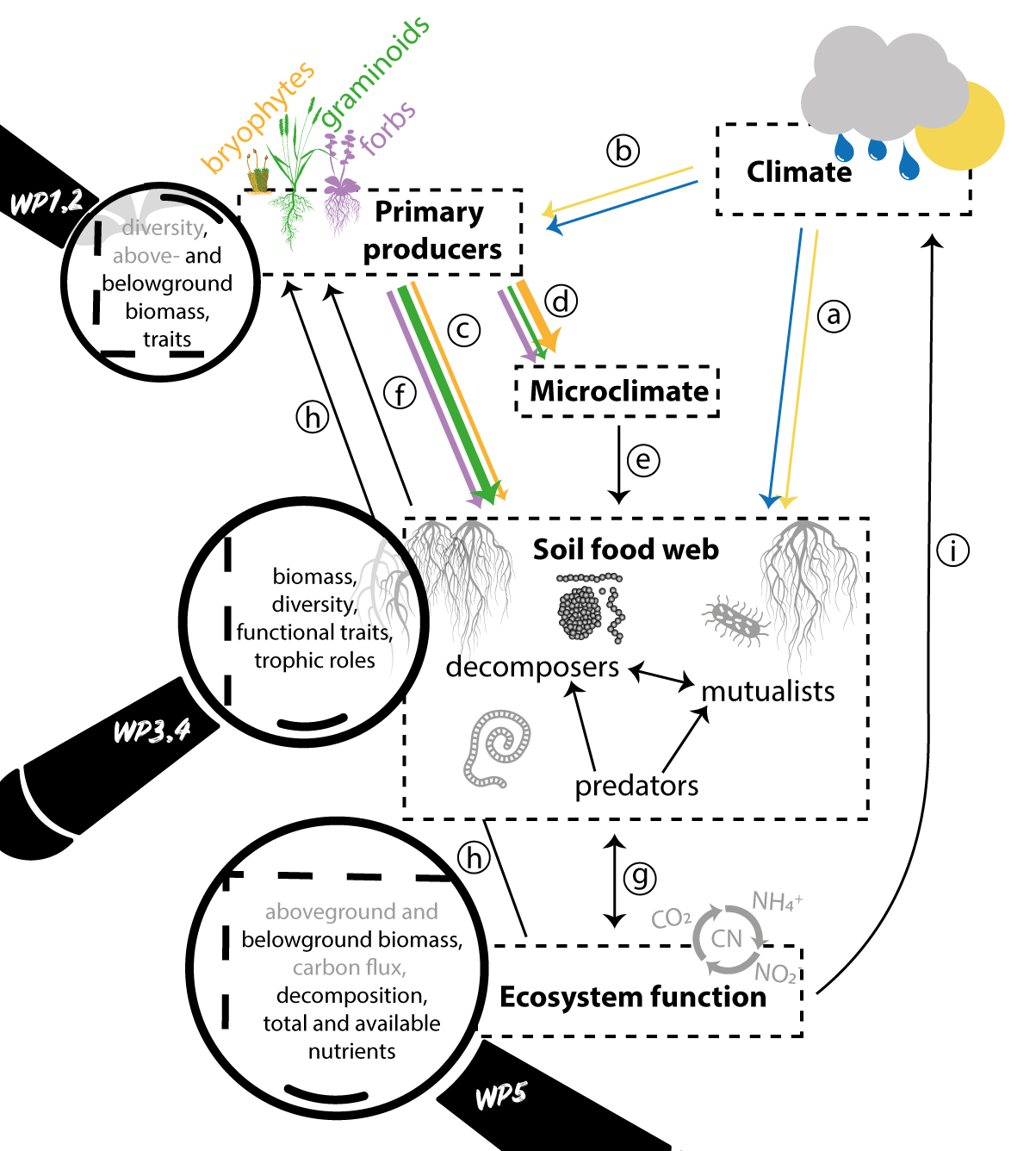
Climate change alters plant and soil communities, as well as processes and interactions in the plant-soil food web. These changes pose threats to biodiversity and key ecosystem functions, such as productivity and carbon and nutrient cycling. To predict how biodiversity and ecosystem functioning will respond to future climatic changes, and how these changes will feed back to the climate system, profound knowledge of climate impacts on underlying ecological responses, processes, mechanisms, and interactions in the plant-soil food web is needed.
FUNDER will assess and disentangle the direct effects of climate from the indirect effects, mediated through biotic interactions, on the diversity and whole-ecosystem functioning of the plant−soil food web. To achieve this, we use a powerful macroecological experimental approach to quantify the impacts of vegetation diversity on interactions and ecosystem functioning across factorial broad-scale temperature and precipitation gradients. This will allow us to gain a holistic understanding of ecosystem responses to climate change, including non-additive effects, context-dependencies across landscapes, compensatory effects and climate mismatches that may lead to disruption of biotic interactions.
Our objectives are to
- Disentangle direct and indirect climate impacts on plants (WP2), soil nematodes and microarthropods (WP3), and soil microbes (WP4), and ecosystem (WP1-4),
- Understand landscape variation and whole-ecosystem consequences of indirect effects, and
- Understand climate feedbacks of the plant-soil food web (WP5).
Our study is conducted across the twelve calcareous grassland experimental sites in the Vestland Climate Grid (VCG), in south-western Norway. The VCG sites were chosen to fit within a climate grid reflecting a crossed design encompassing the major bioclimatic variation in Norway, identified using a combination of topographic maps, geological maps (NGU) and interpolated maps of summer temperature and annual precipitation normals 1960-1990 (100 m resolution gridded data, met.no; see 29 and references therein). The twelve sites are arranged across three temperature levels (alpine, sub-alpine, boreal) replicated across each of four levels of precipitation selected to reflect a difference in mean growing season temperature of ca. 2°C (i.e., the four warmest months of the year) between temperature levels (6.5°C, 8.5°C, 10.5°C) and a difference in mean annual precipitation of 700 mm between precipitation levels (700 mm, 1400 mm, 2100 mm, 2800 mm). The final sites were selected, ensuring that other factors such as grazing regime and history, bedrock, vegetation type and structure, slope and exposure were kept as constant as possible among the selected sites30. Geographical distance between sites is on average 15 km and ranges from 175 km to 650 m.
The functional group removal experiment was designed to examine the impact of aboveground interactions among the major plant functional group, graminoids, forbs and bryophytes, on the performance and functioning of other components of the vegetation and ecosystem. The experiment consists of eight 25×25 cm plots per site and block, with a fully factorial combination of removals of three plant functional groups, with treatments randomized within each block (Figure 1c). The functional groups are abbreviated as follows: G = graminoids (including grasses, sedges and rushes), F = forbs (including herbaceous forbs, pteridophytes, dwarf-shrubs, and small individuals of trees and shrubs), B = Bryophytes (including mosses, liverworts, and hornworts). Note that the species are coded by the functional group into which they were classified in the FunCaB taxon table. The treatments were coded by functional group removed so that FGB = all plants removed, FB = only graminoids remaining, GB = only forbs remaining, GF = only bryophytes remaining, B = graminoids and forbs remaining, F = bryophytes and graminoids remaining, G = bryophytes and forbs remaining, and C = no removal controls. In 2016, four extra control (XC) plots were marked per site for aboveground biomass harvest and ecosystem carbon flux measurements. This sampling regime gave a total of 384 plots, plus the additional 48 controls in 2016.
Functional group removals were done once in 2015 (at peak growing season due to late snowmelt), twice per year in 2016 and 2017 (after the spring growth and at peak growing season) and annually from 2018 - 2022 (at peak growing season) as regrowth had declined (see below) and biannual removals were no longer necessary. At each sampling, all above-ground biomass of the relevant plant functional group was removed from each plot as follows: For each plot, all the above-ground parts of the relevant functional group(s) were removed using scissors and tweezers to cut the plants at the ground layer. Roots and other below-ground parts were not removed, and the non-target vegetation and litter were left intact.
The project description, an overview of all the datasets, and the data dictionaries are in this readme file, available on GitHub. The draft for the data paper is available here. (only available for authors)
The raw and clean datasets from this project are stored and available on OSF.
All R code for the cleaning the raw data is available on GitHub.
| Files or variable | Naming convention | Example |
|---|---|---|
| Project | Project name | FUNDER or FunCaB |
| Datasets | Project_Status_(Experiment)_Response_Year(s).Extension | FUNDER_clean_microbial_community_2022-2023.csv |
| siteID | Unique site ID written out fully | Vikesland, Alrust |
| blockID | Unique block ID, with 3 first letters of siteID and a number (1-4) | Alr1 |
| plotID | Unique plot ID with blockID and treatment | Alr1FGB |
| treatment | Plant functional groups removed, where F = forbs, G = graminoids, B = bryophytes, C = control | FGB, FG, FB, GB, G, F, B, C |
| removal_fg | Removed functional group, where F = forbs, G = graminoids, B = bryophytes. | F, G, B |
| species | Vascular plant taxon names follow for Norway Lid & Lid (Lid J & Lid, 2010). The full taxa is written using genus and species with a blank. | Leontopodium nivale |
| responses | Response variables | cover, biomass, Reco |
Here is the list of valid siteIDs in VCG.
| siteID |
|---|
| Fauske |
| Vikesland |
| Arhelleren |
| Ovstedalen |
| Alrust |
| Hogsete |
| Rambera |
| Veskre |
| Ulvehaugen |
| Lavisdalen |
| Gudmedalen |
| Skjelingahaugen |
Below is code to clean the site names. On the left side are the old names (e.g. Gud) that you want to replace (change to what fits your data). And on the right side are valid names, which will replace the old names (don’t change!).
# code to clean site names
dat |>
mutate(siteID = recode(siteID,
# old name (replace) = valid name (do not change)
'Gud' = "Gudmedalen",
'Lav' = "Lavisdalen",
'Ram' = "Rambera",
'Ulv' = "Ulvehaugen",
'Skj' = "Skjelingahaugen",
'Alr' = "Alrust",
'Arh' = "Arhelleren",
'Fau' = "Fauske",
'Hog' = "Hogsete",
'Ovs' = "Ovstedalen",
'Vik' = "Vikesland",
'Ves' = "Veskre"))This is an overview over all the datasets. They are available on OSF.
| Response | Time period | Level | Project | Filename |
|---|---|---|---|---|
| Site level | ||||
| Coordinates, elevation | - | Site | VCG | e.g. VCG_clean_coordinates.csv |
| Geology, Land-use history | - | Site | VCG | |
| 1) Vegetation | ||||
| Vascular plant species cover | 2015 - 2019, 2022 | Plot | FunCaB | |
| Vascular plant species presence | 2015 - 2019, 2022 | Subplot | FunCaB | |
| Vegetation height | 2015 - 2019, 2022 | Plot | FunCaB | |
| Functional group biomass | 2015 - 2022 | Plot | FunCaB/FUNDER | |
| Total biomass | 2022 | Plot | FUNDER | |
| Reflectance | 2021 | Plot | FunCaB | |
| Root biomass | 2022 | Plot | FUNDER | |
| Root productivity | 2022 | Plot | FUNDER | |
| Root traits | 2022 | Plot | FUNDER | |
| Bryophyte composition | 2022 | Plot | FUNDER | |
| Bryophyte presence? | 2022 | Plot | FUNDER | |
| Bryophyte functional traits | 2022 | Plot | FUNDER | |
| 2) Mesofauna | ||||
| Mycelia production | 2022 | Plot | FUNDER | |
| Mesofauna functional groups and diversity | 2022 | Plot | FUNDER | |
| Mesofauna abundance and biomass | 2022 | Plot | FUNDER | |
| 3) Fungi | ||||
| Fungal functional groups and diversity | 2022 | Plot | FUNDER | |
| 4) Bacteria | ||||
| Bacteria functional groups and diversity | 2022 | Plot | FUNDER | |
| 5) Carbon cycling | ||||
| Ecosystem carbon fluxes | 2015-2018, 2022 | Plot | FunCaB/FUNDER | |
| Litter bag decomposition | 2022 | Plot | FUNDER | |
| Tea bag decomposition | 2022 | Plot | FUNDER | |
| 6) Nutrient cycling | ||||
| C and N stocks | 2022 | Plot | FUNDER | |
| Available nutrients | 2022 | Plot | FUNDER | |
| Soil depth | 2022 | Plot | FUNDER | |
| 7) Climate | ||||
| Soil temperature and moisture | 2022 | Plot | FUNDER | |
| Soil temperature and moisture | 2015-2017 | Plot | FunCaB | |
| Climate | 2009-2022 | Site | VCG |
How to make a data dictionary?
Make data description table
Find the file R/data_dic/data_description.xlsx. Enter all the variables into that table, including variable name, description, unit/treatment level and how measured. If the variables are global for all of Funder, leave TableID blank (e.g. siteID). If the variable is unique for a specific dataset, create a TableID and use it consistently for one specific dataset. Make sure you have described all variables.
Make data dictionary
Find the file R/data_dic/data_dic.R. Add code to import the dataset, e.g. read_csv and give it a name. Then run the function make_data_dic() on your dataset. Check that the function produces the correct data dictionary.
Add data dictionary to readme file
Finally, add the data dictionary below to be displayed in this readme
file. Add a title, and a code chunk using kable() to display the data
dictionary.
knitr::kable(biomass_dic)