Code for converting between MRtrix and DSI Studio. Not a full python package but a proof of concept.
If you ran dwi2fod and got at .mif file containing sh coefficients
you can convert to DSI Studio's fib format if you also have a mask file
Here is a heavily downsampled ABCD-style scan reconstructed with CSD:
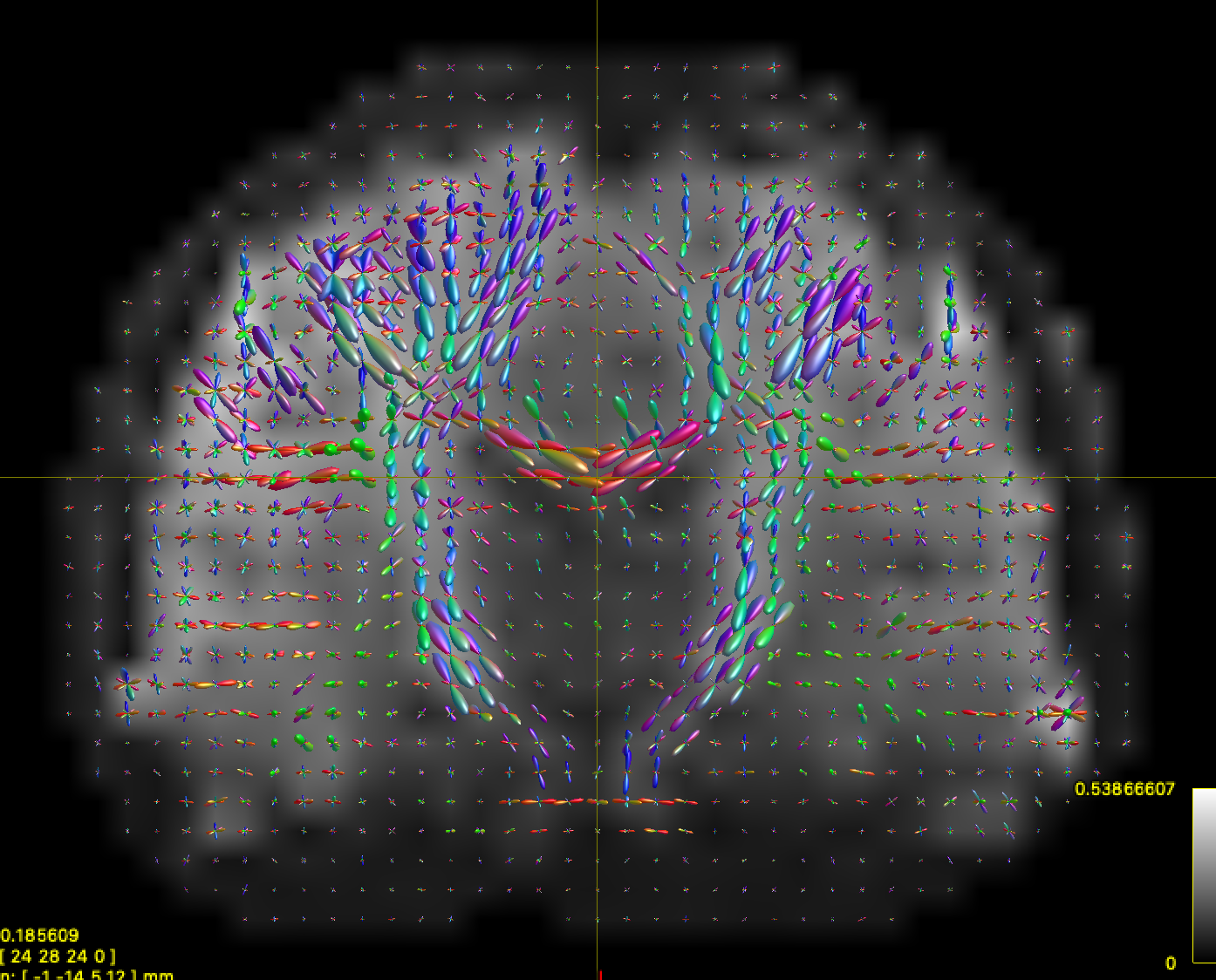
running
from mrtrix_to_dsistudio import mrtrix_to_dsistudio
mrtrix_to_dsistudio("input_sh.mif", "input_sh_mask.nii.gz", "test.fib")results in very nice-looking ODFs in DSI Studio:
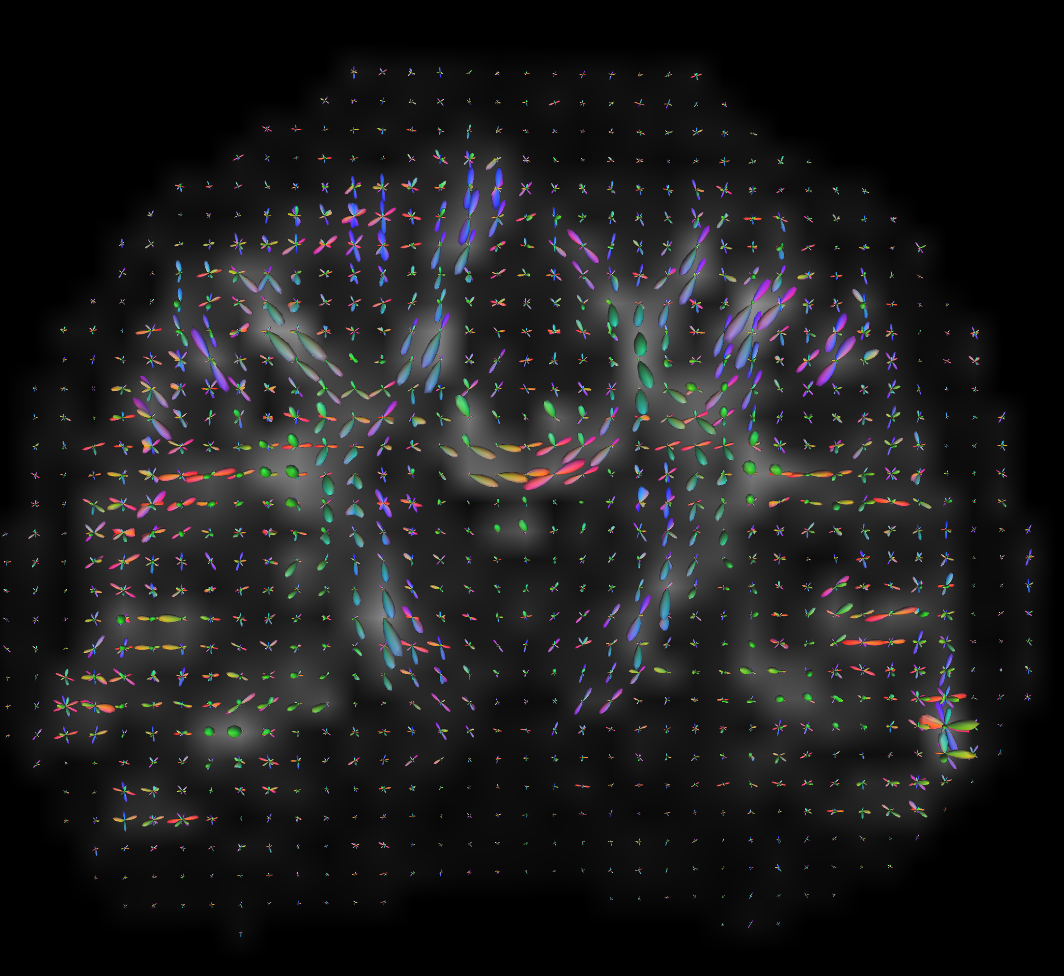
One cool result is that AFD is stored in the fa0, fa1, etc variables.
You can reconstruct in DSI Studio and export to a MRTrix mif file if
you have a mask file in nifti format.
Here is the same ABCD-style data reconstructed in DSI Studio using GQI
with default parameters:
running
from dsistudio_to_mrtrix import dsistudio_to_mrtrix
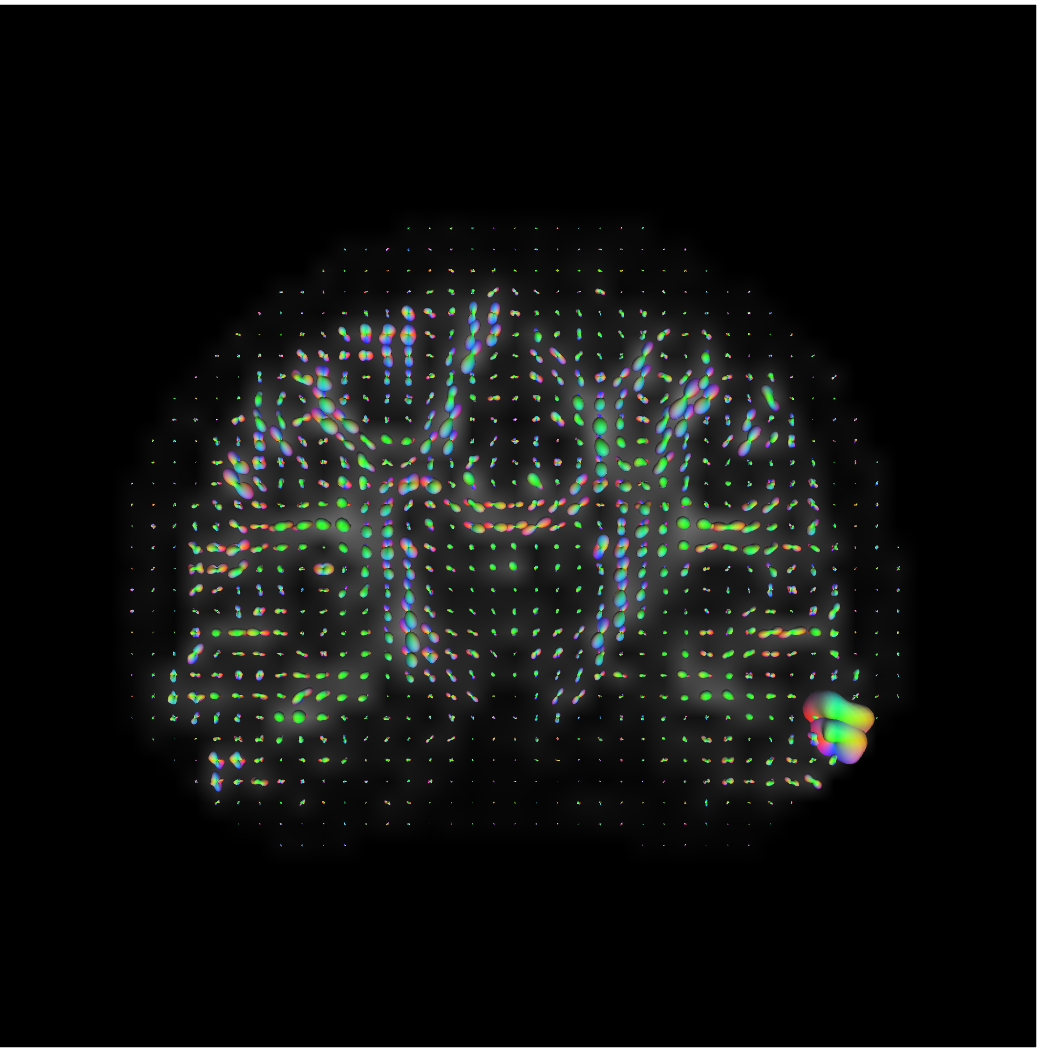
dsistudio_to_mrtrix("dmri.src.odf8.f5rec.bal.csfc.gqi.1.25.fib", "input_sh_mask.nii.gz", "from_dsistudio.mif")results in a blobby, but correct rendering of the GQI ODFs in mrview:
This is actually correct -- DSI Studio subtracts the isotropic component
of each ODF before rendering the glyph. There is no need to do this on
CSD data, so MRTrix doesn't implement that feature. We can tell that
orientation has been preserved by running fod2fixel on the output
of this function:
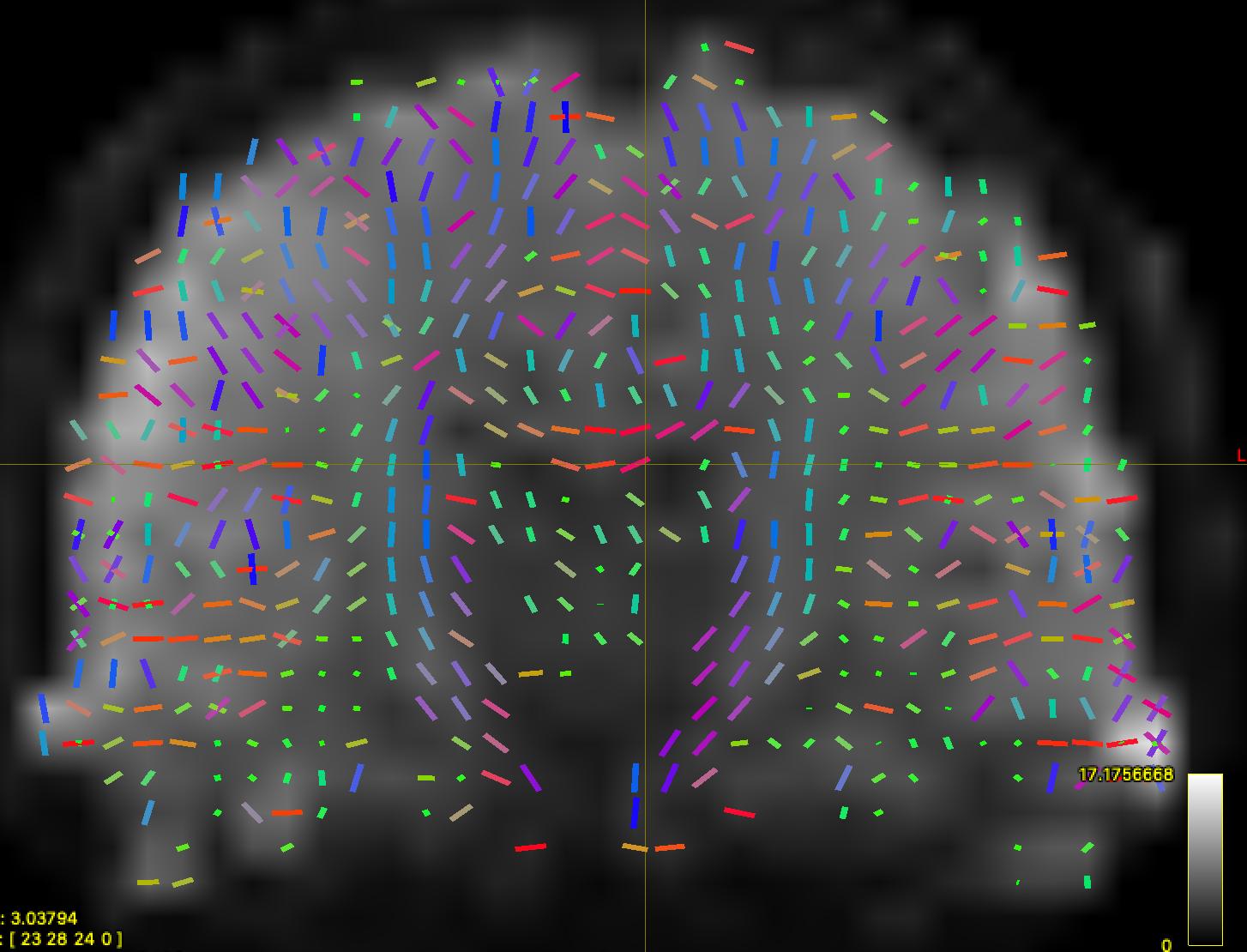
Another cool result is that the AFD here is actually quantitative anisotropy from DSI Studio!