This code performs some of the experiments in
O. Balabanov, M. Beaupere, L. Grigori, V. Lederer. Block subsampled randomized Hadamard transform for low-rank approximation on distributed architectures. View on arXiv.
Given a dense, large, distributed matrix
Furthermore, it also serves the illustrations of my Ph.D thesis.
- Conservation of the inner product through sketching.
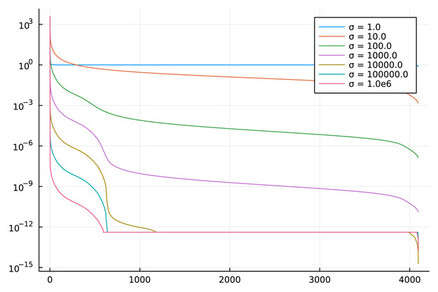
- Obtain singular values from several datasets.
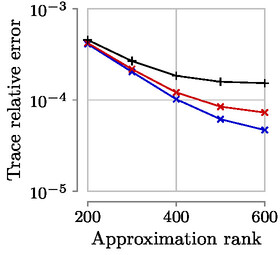
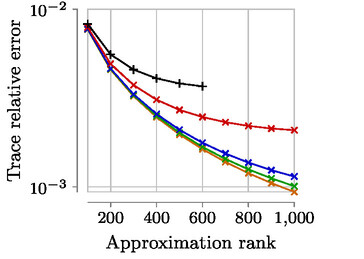
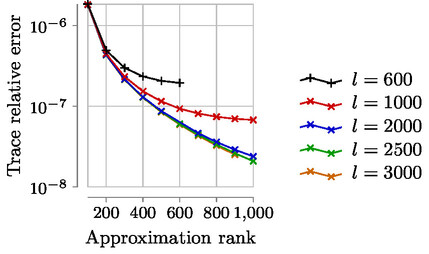
Expected accuracy results (Datasets MNIST, Year with sigma=1000, Year with sigma=10 000):
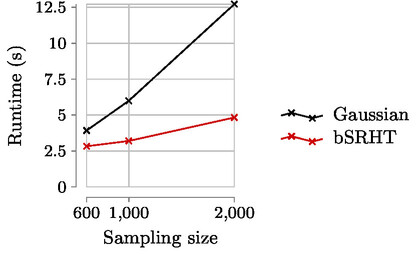
Expected runtime results:
The experiments were done under a Linux environment.
This program requires Julia version 1.7.2 with the following packages
- TimerOutputs.jl
- ClusterManager.jl
- DistributedArrays.jl
- Hadamard.jl.
Download and extract the training and test datasets.
$> wget https://www.csie.ntu.edu.tw/\~cjlin/libsvmtools/datasets/multiclass/mnist.scale.bz2
$> wget https://www.csie.ntu.edu.tw/\~cjlin/libsvmtools/datasets/multiclass/mnist.scale.t.bz2
$> bzip2 -d mnist.scale.bz2
$> bzip2 -d mnist.scale.t.bz2
Concatenate datasets, merging 60 000 records from the training data and 5536 records from the testing data.
The suffix specifying the feature count _780 is required for the program to work correctly.
$> cat mnist.scale > mnist_780
$> head -n 5536 mnist.scale.t >> mnist_780
Or build a datasets of 32 768 rows
$> head -n 32768 mnist.scale >> mnist_780
Load environment variables corresponding to the dataset.
$> source scripts/env/mnist.env
Start Julia
$> ./path/to/julia -p 64
And then start the script to reproduce the timings and the error.
julia> include("scripts/nystrom.jl")
The scripts in scripts/env can help to execute this on a cluster equipped with the Slurm scheduler.
Look at the file timings_p64_kmin200_height65536.csv for the times in milliseconds. Columns are
- The sampling parameter
- The time of block SRHT
- The time of TSQR
- The time of SVD of R
- The time of Gaussian sampling
Only (2) and (5) are given in the submitted paper.
Look at the files nuclear_error_*.csv to get the relative trace error of each run.
This section describes how to reproduces figures from my PhD thesis.
The previous step can be reproduced with other datasets:
Expected result for the MNIST dataset:
Use the scripts/plot_rbs_kernel_svd.jl file and modify the constants:
dataset_filename: Path to dataset.sep: Dataset values separation character.sigmas: Different values of sigma (see RBS kernel expression), each corresponding to a line in the plot.
Use the scripts/innerproducts.jl file. Call the main function with the wanted
dataset (including random) and value for