The recent Learn2Reg challenge shows that single-scale U-Net architectures (e.g. VoxelMorph) with spatial transformer loss, do not generalise well beyond the cranial vault and fall short of state-of-the-art performance for abdominal or intra-patient lung registration.
VoxelMorph++ takes two straightforward steps that greatly reduce this gap in accuracy.
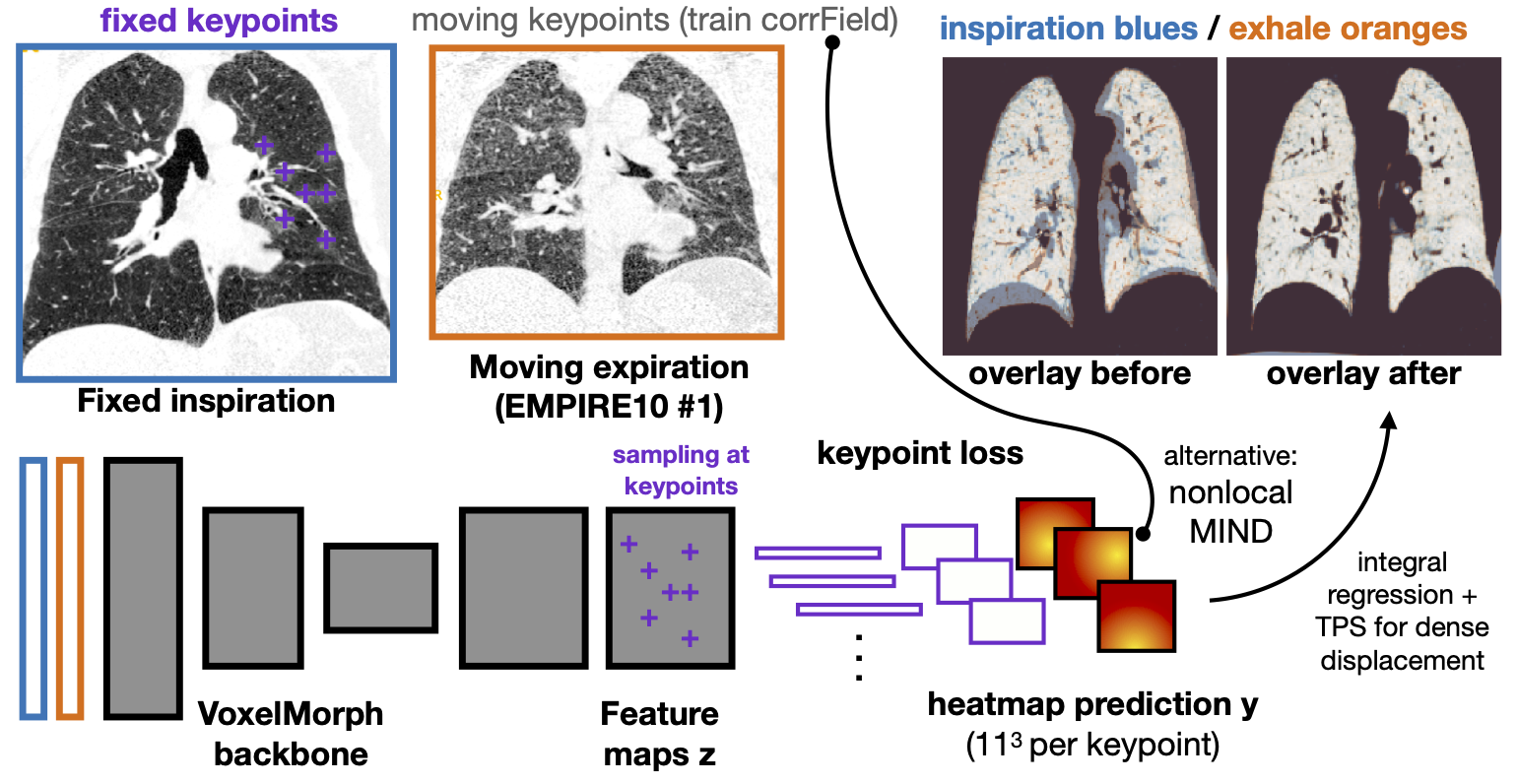
- we employ keypoint self-supervision with a novel heatmap prediction network head
- we replace multiple learned fine-tuning steps by a single instance optimisation with hand-crafted features and the Adam optimiser.
VoxelMorph++ robustly estimates large deformations using the discretised heatmaps and unlike PDD-Net does not require a fully discretised architecture with correlation layer. If no keypoints are available heatmaps can still be used in an unsupervised using only a nonlocal MIND metric.
We outperform VoxelMorph by improving nonlinear alignment by 77% compared to 22% - reaching target registration errors of 2 mm on the DIRlab-COPD dataset. Extending the method to semantic features sets new stat-of-the-art performance of 70% on inter-subject abdominal CT registration. Our network can be trained within 17 minutes on a single RTX A4000 with a carbon footprint of less than 20 grams.
If you find (parts of) this method useful in your work please consider citing:
"Voxelmorph++ going beyond the cranial vault with keypoint supervision and multi-channel instance optimisation." by Heinrich, Mattias P., and Lasse Hansen. In Biomedical Image Registration: 10th International Workshop, WBIR 2022, Munich, Germany, July 10–12, 2022, Proceedings, pp. 85-95. Cham: Springer International Publishing, 2022.
Our core new element is the heatmap prediction network head, which requires a number of 1D feature vectors sampled at keypoints as input and predicts a discretised heatmap of shape 11x11x11:
heatmap = nn.Sequential(nn.ConvTranspose3d(64,16,7),nn.InstanceNorm3d(16),nn.ReLU(),nn.Conv3d(16,32,3,padding=1),nn.InstanceNorm3d(32),nn.ReLU(), nn.Conv3d(32,32,3,padding=1),nn.Upsample(size=(11,11,11),mode='trilinear'),nn.Conv3d(32,32,3,padding=1),nn.InstanceNorm3d(64),nn.ReLU(), nn.Conv3d(32,32,3,padding=1),nn.InstanceNorm3d(32),nn.ReLU(),nn.Conv3d(32,16,3,padding=1),nn.InstanceNorm3d(16),nn.ReLU(),nn.Conv3d(16,1,3,padding=1))
It is composed of one transpose convolution, a trilinear upsampling and 6 additional 3D convolutions. Let output be the U-Net features, keypts_xyz the sampling locations and mesh an affine grid with appropriate capture range setting. We can easily estimate a sparse 3D displacement field as follows:
sampled = F.grid_sample(output,keypts_xyz.cuda().view(1,-1,1,1,3),mode='bilinear')
soft_pred = torch.softmax(heatmap(sampled.permute(2,1,0,3,4)).view(-1,11**3,1),1)
pred_xyz = torch.sum(soft_pred*mesh.view(1,11**3,3),1)
When using a nonlocal MIND or nnUNet loss we use soft_pred as discrete displacement probabilities. pred_xyz can be extrapolated to a dense field using thin-plate-splines.
We slightly adapt the basic Voxelmorph U-Net as a backbone baseline, adding some more convolutions and feature channels
unet_half_res=True; bidir=False; use_probs=False; int_steps=7; int_downsize=2
def default_unet_features():
nb_features = [
[32, 48, 48, 64],[64, 48, 48, 48, 48, 32, 64]]
return nb_features
class ConvBlock(nn.Module):
def __init__(self, ndims, in_channels, out_channels, stride=1):
super().__init__()
Conv = getattr(nn, 'Conv%dd' % ndims)
self.main = Conv(in_channels, out_channels, 3, stride, 1)
self.norm = nn.InstanceNorm3d(out_channels); self.activation = nn.ReLU()
self.main2 = Conv(out_channels, out_channels, 1, stride,0)
self.norm2 = nn.InstanceNorm3d(out_channels); self.activation2 = nn.ReLU()
def forward(self, x):
out = self.activation(self.norm(self.main(x)))
out = self.activation2(self.norm2(self.main2(out)))
return out
unet_model = Unet(ConvBlock,inshape,infeats=2,nb_features=nb_unet_features,nb_levels=nb_unet_levels,\
feat_mult=unet_feat_mult,nb_conv_per_level=nb_unet_conv_per_level,half_res=unet_half_res,)
The model has 901'888 trainable parameters and produces 64 output features.
The keypoint loss is a simple mean-squared error penalty
loss = nn.MSELoss()(pred_xyz,disp_gt[idx])
To run this code you need scans resampled to 1.75 x 1.25 x 1.75 mm (exhale) and 1.75 x 1.00 x 1.25 mm (inspiration) with dimensions 192 x 192 x 208 voxels. For each of them a binary lung mask should be provided (we will soon make available a segmentation network that can automate both steps). Pre-trained models are available under data. To reproduce our DIRlab-COPD evaluation experiment simple run the following (which will automatically select the correct cross-validation folds):
folds=(0 4 5 4 3 5 3 2 1 2 1)
for i in {01..10}; do python inference_voxelmorph_plusplus.py -F COPDgene/case_${i}_exp.nii.gz -M COPDgene/case_${i}_insp.nii.gz -f COPDgene/case_${i}_exp_mask.nii.gz -m COPDgene/case_${i}_insp_mask.nii.gz-n l2r_lung_ct/repeat_l2r_voxelmorph_heatmap_keypoint_fold${folds[10#$i]}.pth -d COPDgene/disp_${i}.pth -c ${i}; done
There are options -d and -w to write out displacement fields and warped moving scan to files. If neither is supplied an coronal overlay png is created.