This is a python script to evaluate the performance of a classification model. The script is designed to help a fellow Data Scientist asses the performance of a model and potentially debug / or assure their model’s performance.
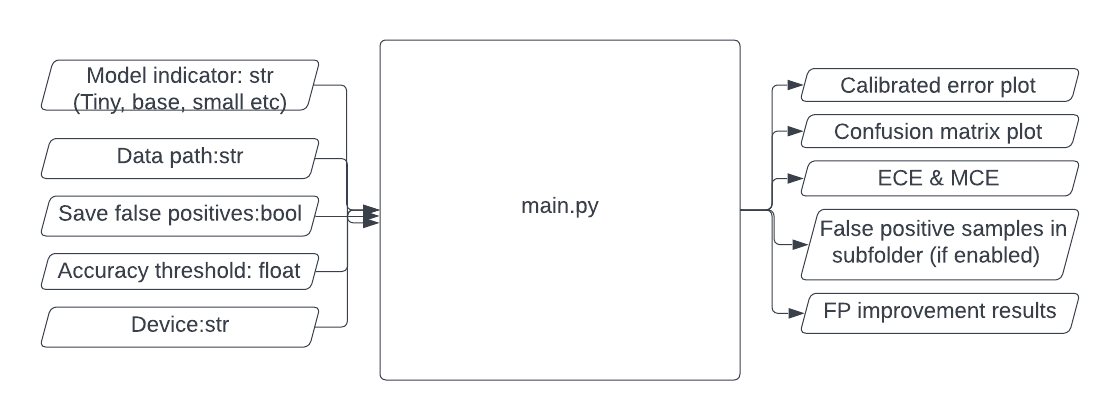 The script main.py takes in 5 parameters:
The script main.py takes in 5 parameters:
- model - optional: tiny / small / base / large. Default to tiny.
- data - default to data/mnist/test.
- save_fp - decide if to save false positive samples into sub-folder results/false_positives.
- acc_thresh - default to 0. If > 0, the early stopping procedure will be activated.
- device - default to 'cuda:0'.
Script main.py gives the following output:
- Plots for calibrated errors and confusion matrix are saved in results.
- ECE and MCE values are printed on-screen.
- False positive samples are stored in results/false_positives.
- False positive improvement results are printed on-screen. Print nothing if false_positives folder does not exist or is empty.
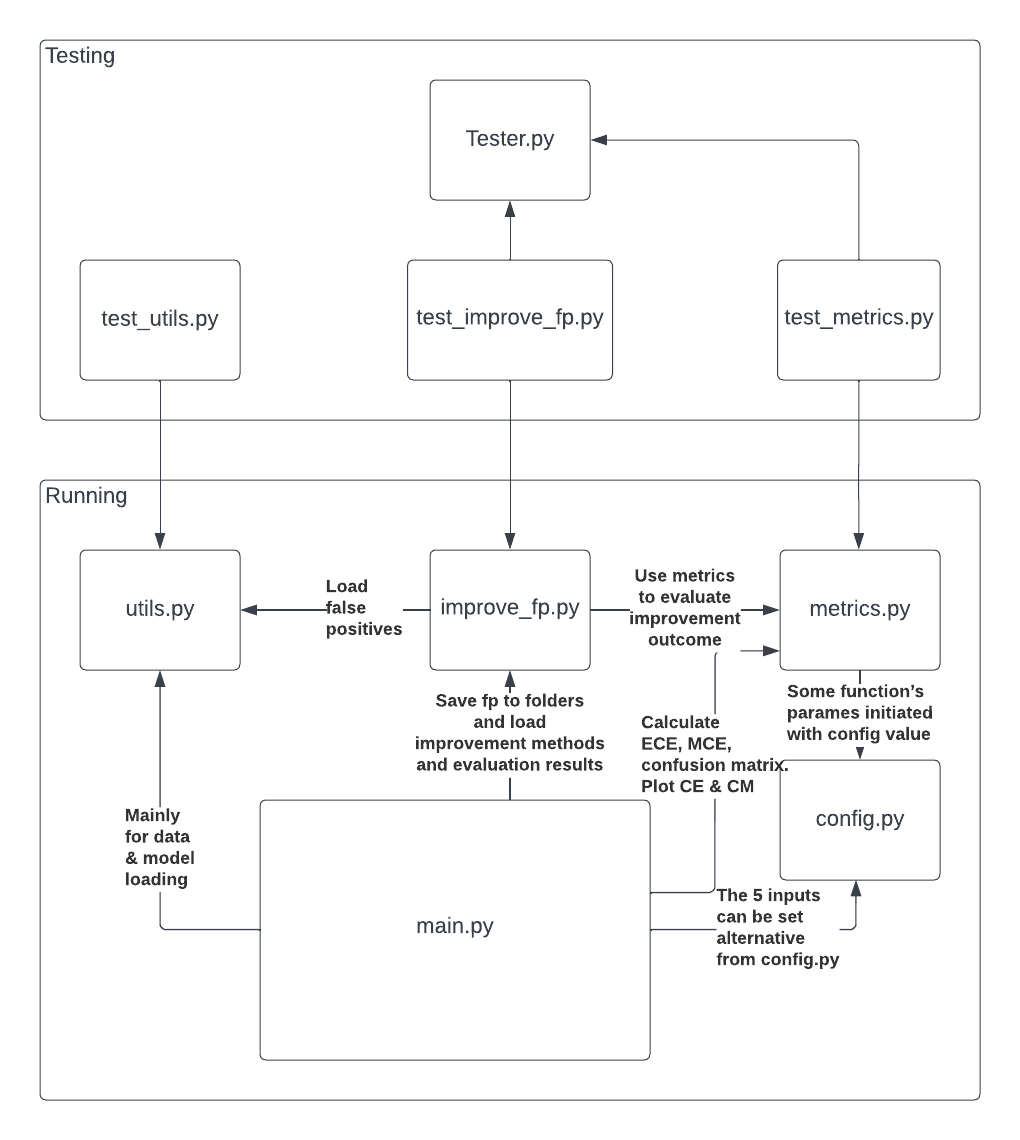 The script components can be divided into two blocks - running and testing. The order of import and usages are demonstrated in above diagram.
The script components can be divided into two blocks - running and testing. The order of import and usages are demonstrated in above diagram.
Note: The 5 input params can be set alternatively with config.py as well as other hyper parameters.
Set parameter acc_thresh to value above 0 to enable this behavior.
- UserWarning: Model accuracy below set threshold. Terminating evaluation now...Assume dataloader randomly shuffles, if average accuracy is lower than set threshold, the script will terminate batch iteration and carry on with the rest of procedure (return metric values, save plots, save false positives and improve for batches so far).
In case of any changes to the scripts or using scripts on new inputs, tests against functions are created to quickly examine the validity of function or compatibility between new input and the function to save debugging efforts. Testing scripts are saved in testing sub-folder.
The tests are to assert matching of set input-out pairs that are designed to handle different case scenarios (except plot tests which relies on manual check of plots). For example, 2 cases are covered for MCE, ECE tests - when some bins are of 0 count vs. all bins are of 0 count:
assert round(float(ece), 3) == 0.192, "wrong ECE for normal CE"
assert round(float(mce), 3) == 0.390, "wrong mce for normal CE"
assert get_mce(ce) == -0.1, "wrong MCE for all non-existent CE"
assert get_ece(ce_b, batch_size) == -0.1, "wrong ECE for all non-existent CE"For confusion matrix test, 3 scenarios are covered - when some prediction is correct / all are correct / prediction and label set is empty:
cm = get_confusion_matrix(y_pred, y_true, n_class=class_size)
assert cm.tolist() == [[2, 2, 1],
[0, 2, 1],
[1, 0, 1]], "wrong confusion matrix computation"
test_plot_cm(cm, "cm_normal")
cm = get_confusion_matrix(y_pred, y_pred, n_class=class_size)
assert cm.tolist() == [[3, 0, 0],
[0, 4, 0],
[0, 0, 3]], "wrong confusion matrix computation"
test_plot_cm(cm, "cm_all_correct")
cm = get_confusion_matrix(torch.tensor([]), torch.tensor([]), n_class=class_size)
assert cm.tolist() == torch.zeros(class_size, class_size).int().tolist(), \
"wrong confusion matrix computation"
test_plot_cm(cm, "cm_empty")Note: Need to pass all tests before branch merge.
The idea is to reduce data dimensionality and perform clustering on the lower D dataset.
This script uses T-sne for d-reduction and K-means for clustering.
Accuracy increases around 10 times for both MNIST and CIFAR10 datasets after improving over false positives. Change parameter n_cluster from config.py to receive different outcomes. n_cluster is default to 256.
For example, for ConvNext tiny + MNIST:
ECE value: 0.6664446592330933
MCE value: 0.6827665567398071
Accuracy: 0.06425222009420395
Plots saved at results
Improving the false positives with model from improve_fp.py ...
Processing with CPU. This could take a while ...
Accuracy after improvement: 0.7463933825492859
-
CovNext model doesn't work with MNIST dataset with a padding lower than 2. It returns this error:
- RuntimeError: Calculated padded input size per channel: (1 x 1). Kernel size: (2 x 2). Kernel size can't be greater than actual input sizeSetting padding to 2 will fix it, but I haven't figured out why due to time limitation and being not familiar with ConvNext model. Adjusting padding automatically will generalize better to wider dataset.
-
The current early stopping strategy relies on accuracy alone. This could be changed to taking {metric: threshold}, letting users define their own multi-threshold. Need to include more metrics and write {metric_name: metric_function} lookup for this to happen.
-
For better evaluation and debugging potential, including more metrics and visualization of metrics will definitely improve usability. For example, for classification task, precision, recall, f1, accuracy, ROC and AUC can be included.
-
Currently, the evaluation outcomes are saved locally, meaning that rerun scripts unless change save path will write over past records. Integration with database, submitting session-id and metric values via database API will allow safer storage and past session lookup, and for visual comparison among sessions with self-made or out-of-shelf tools like tensorboard.
Install the required packages with pip to get started.
- pip
pip install -r requirements.txt
Can simply run main.py with parameters readily defined in config.py:
python3 main.py
Otherwise, please define them with commandline:
python3 main.py -d dataset/mnist/test -m base -t 0.2 -s true --device cpu
To run individual script:
python3 test_metrics.py
To run all:
sh run.sh
The testing plots and false positives are saved in testing/vis and testing/results/false_positives
