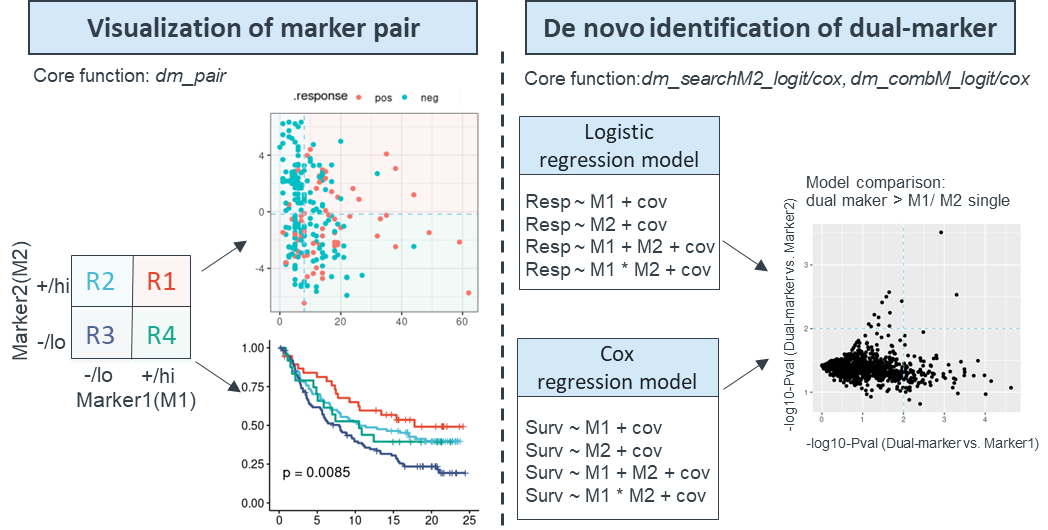
dualmarker
Dualmarker is designed for data exploration and hypothesis generation
for dual biomarkers. It provides intuitive visualizations and extensive
assessment of two marker combinations using logistic regression model
for binary outcome (response analysis) and Cox regression for
time-to-event outcome (survival analysis).
It performs dual marker analysis via two distinct modules:
- One module is evaluation of specific biomarker pair through
dm_pair function, which comprehensively reveals the correlation among
two markers, response and survival using over 14 sub-plots,such as
boxplots, scatterplots, ROCs, and Kaplan-Meier plots.
- Another module is de-novo identification and prioritization of
marker2 among candidate markers in combination with known marker1 to
predict response and survival through dm_searchM2_cox and
dm_searchM2_logit function, its expansion version works for all
biomarker combination to prioritize the most significant pair through
dm_combM_cox and dm_combM_logit function.
It is applicable for both response and survival analyses and compatible
with both continuous and categorical variables. This figure illustrates
the framework of this package.
Installation
Install the latest developmental version from GitHub as follow:
if(!require(devtools)) install.packages("devtools")
devtools::install_github("maxiaopeng/dualmarker")Key functions
Plenty of commands and functions are wrapped into five main functions: dm_pair for visualization and statistics of given dual marker pairs, dm_searchM2_cox and dm_searchM2_logit for de novo identification of novel marker2 to combine with marker1 using Cox survival model and logistic regression model; dm_combM_cox and dm_combM_logit for de novo identification of marker pairs among all dual marker combinations.
dm_pair
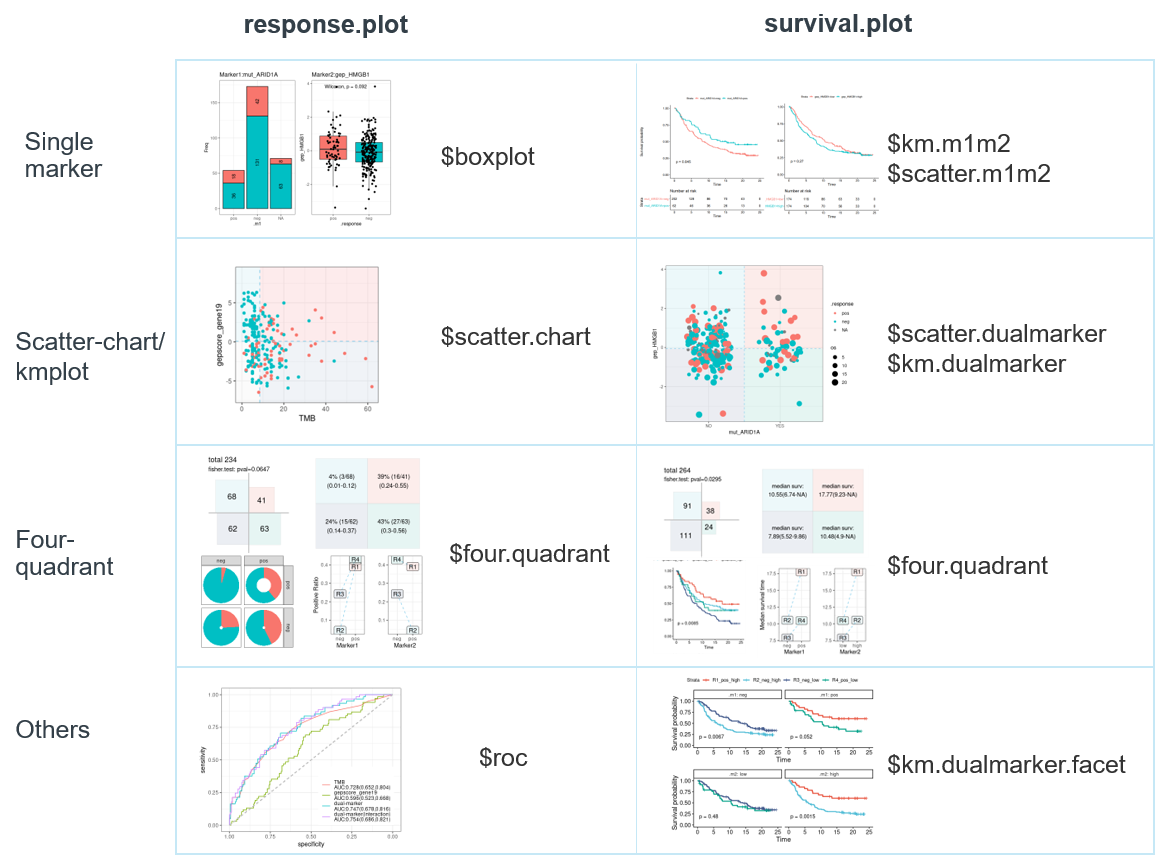
dm_pair is the main function for visualization and statistics of one pair of specific biomarkers. It takes the marker1, marker2, covariates (optional), binary outcome(response) or time-to-event outcome(survival) variable as input, and returns list of comprehensive plots (in response.plot, survival.plot object) and statistic results (in response.stats, survival.stats object) from logistic regression for binary outcome(response) and Cox regression for time-to-event outcome(survival) . The result is a list object with the following structure:
- response.plot
- boxplot
- scatter.chart
- four.quadrant
- roc
- response.stats
- logit
- four.quadrant
- param
- stats
- survival.plot
- km.m1m2
- scatter.m1m2
- km.dualmarker
- km.dualmarker.facet
- scatter.dualmarker
- four.quadrant
- survival.stats
- cox
- four.quadrant
- param
- stats
All visualizations are shown in this overall plot
dm_searchM2_cox/logit
The main functions for do novo identification of marker2 from candidates (m2.candidates) to combine with marker1. It takes marker1, and m2.candidates, covariates (optional), binary outcome(response) or time-to-event outcome(survival) variables as input and returns the statistical result of logistic or Cox regression model. Four regression models are built using single marker and dual marker w/ or w/o interaction term:
- model1: Surv/Resp ~ marker1 + covariates, labeled as ‘SM1’ for short
- model2: Surv/Resp ~ marker2 + covariates , labeled as ‘SM2’ for short
- model3: Surv/Resp ~ marker1 + marker2 + covariates, labeled as ‘DM’ for short, i.e. dual-marker
- model4: Surv/Resp ~ marker1 * marker2 (with interaction term) + covariates, labeled as ‘DMI’ for short, i.e. dual-marker with interaction
Model comparison is performed between dual-marker and single-marker
models by Likihood ratio test(LRT) using anova function.
dm_searchM2_topPlot can facilitate the glance of top candidate
marker2s for both logistic and Cox regression.
The statistics of regression include the basic information of marker1,
marker2, covariates, response, time, event; the estimate and p-values
from 4 models, ROC/AUC(for binary outcome), concordant probability
estimate(CPE, for survival analysis), AIC, p-values of model
comparisons. Users can filter and get interesting dual-marker pairs
using the statistics and model performance metrics. Here is an example
of logistic regression:
| colnames | example values | description |
|---|---|---|
| response | binaryResponse | response variable |
| response_pos | CR, PR | positive response |
| response_neg | SD, PD | negative response |
| m1 | TMB | marker1 variable |
| m2 | gepscore_TGFb | marker2 variable |
| covariates | "" | covariates variable |
| cutpoint_m1 | NA | cutpoint if m1 is numeric |
| cutpoint_m2 | NA | cutpoint if m2 is numeric |
| m1_cat_pos | "" | positive values if m1 is categorical |
| m1_cat_neg | "" | negative values if m1 is categorical |
| m2_cat_pos | "" | positive values if m2 is categorical |
| m2_cat_neg | "" | negative values if m2 is categorical |
| SM1_m1_estimate | 0.09963989 | estimate of marker1 in SM1 model |
| SM1_m1_p.value | 1.395883e-06 | pvalue of marker1 in SM1 model |
| SM2_m2_estimate | -0.1199803 | estimate of marker2 in SM2 model |
| SM2_m2_p.value | 0.02516686 | pvalue of marker2 in SM2 model |
| DM_m1_estimate | 0.09663392 | estimate of marker1 in DM model |
| DM_m1_p.value | 2.582342e-06 | pvalue of marker1 in DM model |
| DM_m2_estimate | -0.08748326 | estimate of marker2 in DM model |
| DM_m2_p.value | 0.135587 | pvalue of marker2 in DM model |
| DMI_m1_estimate | 0.119926 | estimate of marker1 in DMI model |
| DMI_m1_p.value | 1.162081e-06 | pvalue of marker1 in DMI model |
| DMI_m2_estimate | -0.2967303 | estimate of marker2 in DMI model |
| DMI_m2_p.value | 0.003635338 | pvalue of marker2 in DMI model |
| DMI_m1:m2_estimate | 0.0182465 | estimate of interaction m1:m2 |
| DMI_m1:m2_p.value | 0.007925505 | pvalue of interaction m1:m2 |
| SM1_auc | 0.728 | AUC of SM1 model |
| SM2_auc | 0.591 | AUC of SM2 model |
| DM_auc | 0.747 | AUC of DM model |
| DMI_auc | 0.754 | AUC of DMI model |
| SM1_AIC | 239.2481 | AIC of SM1 model |
| SM2_AIC | 267.3176 | AIC of SM2 model |
| DM_AIC | 238.98 | AIC of DM model |
| DMI_AIC | 233.9904 | AIC of DMI model |
| pval_SM1_vs_NULL | 7.993656e-09 | pvalue of SM1-vs-NULL model |
| pval_SM2_vs_NULL | 0.02249407 | pvalue of SM2-vs-NULL model |
| pval_SM1_vs_DM | 0.1320594 | pvalue of SM1-vs-DM model |
| pval_SM2_vs_DM | 3.630161e-08 | pvalue of SM2-vs-DM model |
| pval_SM1_vs_DMI | 0.009765815 | pvalue of SM1-vs-DMI model |
| pval_SM2_vs_DMI | 7.84323e-09 | pvalue of SM2-vs-DMI model |
dm_combM_cox/logit
two functions for de novo identification of biomarker pairs from all dual-marker combinations. They use the similar input and get same output as dm_searchM2_cox/logit.
dataset
We demonstrate the package using
Imvigor210
biomarker data. This dataset includes the baseline characterization of
PDL1 protein level(IHC), gene expression profiling(GEP) and mutations on
348 advanced UC patients as well as response and Overall survival(OS)
data treated by Atezolizumab.
The demographic info, clinical efficacy and biomarker data is stored in
clin_bmk_IMvigor210 dataframe, with gene expression variables
containing ‘gep_’ prefix, gene signature score variables containing
‘gepscore_’ prefix and mutation variables containing ‘mut_’ prefix.
The GEP data and gene signature score is processed according to the
IMvigor210CoreBiologies package and gene signature score is calculated
using hallmark genesets from MsigDBv7.0 as well as signatures from the
IMvigor210CoreBiologies package.
library(dualmarker)
library(stringr)
library(dplyr)Example1: marker pair of TMB + TGF-beta signature
Here we demonstrate the binary outcome (drug response) analysis of TMB + TGF-beta gene signature(gepscore_TGFb.19gene) using dm_pair function. This pair of biomarker is studied in Nature.2018 Feb 22;554(7693):544-548. The response variable should be dichotomized by setting response.pos and response.neg values.
res.pair <- dm_pair(
data = clin_bmk_IMvigor210,
# response info
response = "binaryResponse",
response.pos = "CR/PR",
response.neg = "SD/PD",
label.response.pos = "R",
label.response.neg = "NR",
# marker1
marker1 = "TMB",
m1.num.cut = "median", # default median cut for continuous variable
label.m1 = "TMB",
label.m1.pos = "TMB_hi",
label.m1.neg = "TMB_lo",
# marker2
marker2 = "gepscore_TGFb.19gene",
m2.num.cut = "median",
label.m2 = "TGF_beta",
label.m2.pos = "TGFb_hi",
label.m2.neg = "TGFb_lo",
# others
na.rm.response = F, # show NA for response variable
na.rm.marker = F, # show NA from marker variable
# palette
palette.4quadrant = "default",
palette.other = "default"
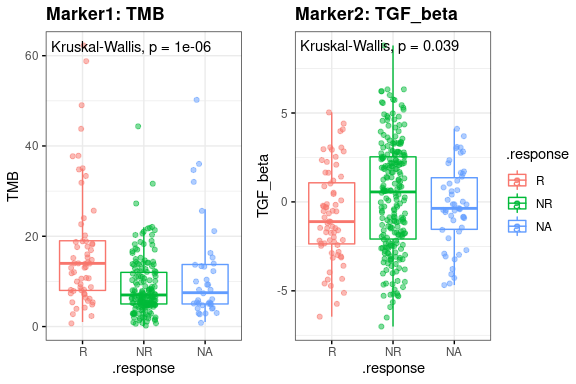
)- plot-1: [response analysis] Single marker
The correlation between single marker and drug response is shown in boxplot, labeled with p-values of Wilcoxon test between positive and negative outcome(response).
res.pair$response.plot$boxplot- plot-2: [response analysis] Scatter chart
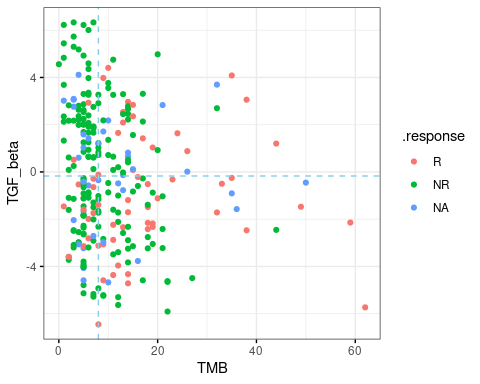
The correlation between marker1, maker2 and drug response is shown in scatter-chart with color indicating response status. If marker1 and/or marker2 is categorical, the jitter plot will be shown.
res.pair$response.plot$scatter.chart- plot-3: [response analysis] Four-quadrant chart
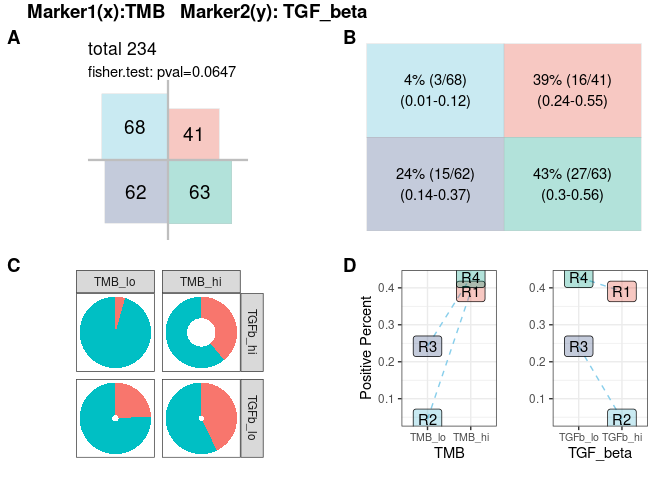
Samples are split into four groups/quadrants(R1-R4), according to cutoffs for continuous markers, default using ‘median’. The independence of each group/quadrant is tested by Fisher exact test. Response rate, sample size and confidence interval are shown in matrix, doughnut chart and line chart. For the doughnut chart, response rate is corresponding to red arc fraction and sample size to width of ring, and the line chart reveals the response rate and potential statistical interaction from two markers if lines are crossed.
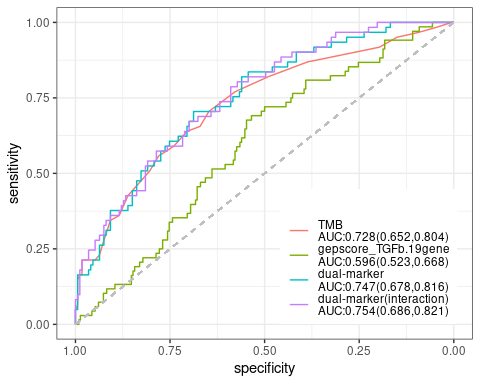
res.pair$response.plot$four.quadrant- plot-4: [response analysis] ROC curve
The single and dual marker prediction of drug response is also shown on ROC curve. Logistics regression model is applied w/ or w/o interaction term between two biomarkers. AUC value and its confidence interval is also drawn on the plot
res.pair$response.plot$roc-
plot-5: [survival analysis] survival plot of dual marker
not available here, see Example2 -
plot-6: [survival analysis] survival plot of dual marker
not available here, see Example2 -
plot-7: [survival analysis] Four-quadrant chart
not available here, see Example2 -
stats-1: [response analysis] Four-quadrant statistics
(response.stats)four.quadrant contains four-quadrant statistics of response, in the following 2 objects:- param contains the information of marker1,marker2, et al.
- stats contains the sample number, response rate and its confidence interval in each quadrant, R1-R4
res.4q <- res.pair$response.stats$four.quadrant
res.4q$param
#> # A tibble: 1 x 7
#> response response.pos response.neg m1 m2 cutpoint.m1 cutpoint.m2
#> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl>
#> 1 binaryResp… CR/PR SD/PD TMB gepscore_… 8 -0.172
res.4q$stats
#> # A tibble: 4 x 9
#> region .m1.level .m2.level n.total n.pos n.neg pct.pos pos.lower95 pos.upper95
#> <chr> <chr> <chr> <int> <int> <int> <dbl> <dbl> <dbl>
#> 1 R1 pos pos 41 16 25 0.390 0.242 0.555
#> 2 R2 neg pos 68 3 65 0.0441 0.00919 0.124
#> 3 R3 neg neg 62 15 47 0.242 0.142 0.367
#> 4 R4 pos neg 63 27 36 0.429 0.305 0.560-
stats-2: [response analysis] Logistic regression result
Four logistic regression models are built. Model comparison between dual-marker and single-marker models is performed by Likelihood ratio test(LRT) using anova function::- model1: Resp ~ marker1 + covariates, labeled as ‘M1’ for short
- model2: Resp ~ marker2 + covariates, labeled as ‘M2’ for short
- model3: Resp ~ marker1 + marker2 + covariates, labeled as ‘MD’ for short, i.e. dual-marker
- model4: Resp ~ marker1 * marker2 + covariates (with interaction term), labeled as ‘MDI’ for short, i.e. dual-marker with interaction
Logistic regression models return the following values:
- basic parameters: time, event, m1(marker1), m2(marker2), cut point of m1/m2 if m1/m2 are continuous, positive/negative values if m1/m2 is categorical
- logistic regression parameters: estimate(weight) and p-value(Wald test) of each predictive variable in model1(SM1), model2(SM2), model3(DM), model4(DMI). ‘MDI_.m1:.m2_estimate’ and ‘MDI_m1:m2_pval’ is estimate and p-value of the interaction term of marker1 and marker2
- AIC: AIC of SM1,SM2,DM and DMI models
- model comparison: p-values of Likelihood Ratio Test(LRT) for SM1-vs-NULL(null model, no marker), SM2-vs-NULL, SM1-vs-DM, SM2-vs-DM, SM1-vs-DMI, SM2-vs-DMI
dplyr::glimpse(res.pair$response.stats$logit)
#> Rows: 1
#> Columns: 40
#> $ response <chr> "binaryResponse"
#> $ response_pos <chr> "CR/PR"
#> $ response_neg <chr> "SD/PD"
#> $ m1 <chr> "TMB"
#> $ m2 <chr> "gepscore_TGFb.19gene"
#> $ covariates <chr> ""
#> $ cutpoint_m1 <lgl> NA
#> $ cutpoint_m2 <lgl> NA
#> $ m1_cat_pos <chr> ""
#> $ m1_cat_neg <chr> ""
#> $ m2_cat_pos <chr> ""
#> $ m2_cat_neg <chr> ""
#> $ SM1_m1_estimate <dbl> 0.09963989
#> $ SM1_m1_p.value <dbl> 1.395883e-06
#> $ SM2_m2_estimate <dbl> -0.1199803
#> $ SM2_m2_p.value <dbl> 0.02516686
#> $ DM_m1_estimate <dbl> 0.09663392
#> $ DM_m1_p.value <dbl> 2.582342e-06
#> $ DM_m2_estimate <dbl> -0.08748326
#> $ DM_m2_p.value <dbl> 0.135587
#> $ DMI_m1_estimate <dbl> 0.119926
#> $ DMI_m1_p.value <dbl> 1.162081e-06
#> $ DMI_m2_estimate <dbl> -0.2967303
#> $ DMI_m2_p.value <dbl> 0.003635338
#> $ `DMI_m1:m2_estimate` <dbl> 0.0182465
#> $ `DMI_m1:m2_p.value` <dbl> 0.007925505
#> $ SM1_auc <dbl> 0.728
#> $ SM2_auc <dbl> 0.591
#> $ DM_auc <dbl> 0.747
#> $ DMI_auc <dbl> 0.754
#> $ SM1_AIC <dbl> 239.2481
#> $ SM2_AIC <dbl> 267.3176
#> $ DM_AIC <dbl> 238.98
#> $ DMI_AIC <dbl> 233.9904
#> $ pval_SM1_vs_NULL <dbl> 7.993656e-09
#> $ pval_SM2_vs_NULL <dbl> 0.02249407
#> $ pval_SM1_vs_DM <dbl> 0.1320594
#> $ pval_SM2_vs_DM <dbl> 3.630161e-08
#> $ pval_SM1_vs_DMI <dbl> 0.009765815
#> $ pval_SM2_vs_DMI <dbl> 7.84323e-09-
stats-3: [survival analysis] Four-quadrant statistics
not available here, see Example2 -
stats-4: [survival analysis] Cox regression result
not available here, see Example2
Example2: marker pair of ARID1A mutation + CXCL13 expression
Here we demonstrate the visualization of CXCL13 expression and ARID1A mutation , this biomarker pair is studied by Sci Transl Med. 2020 Jun 17;12(548):eabc4220, we showed the same result here.
res.pair <- dm_pair(
data = clin_bmk_IMvigor210,
# response (optional)
response = "binaryResponse",
response.pos = "CR/PR",
response.neg = "SD/PD",
label.response.pos = "R",
label.response.neg = "NR",
# survival info
time = "os",
event = "censOS",
# marker1
marker1 = "mut_ARID1A",
label.m1 = "ARID1A-mut",
m1.cat.pos = "YES",
m1.cat.neg = "NO",
label.m1.pos = "Mut",
label.m1.neg = "Wt",
# marker2
marker2 = "gep_CXCL13",
m2.num.cut = "median",
label.m2 = "CXCL13 expression",
label.m2.pos = "Hi",
label.m2.neg = "Lo",
# palette
palette.4quadrant = "jco", # color scheme for four-quadrants
palette.other = "jco", # color scheme for others
# others
na.rm.response = T, # show NA in response variable
na.rm.marker = T # show NA in biomarker variable: mut_ARID1A, gep_CXCL13
)-
plot-1: [response analysis] Single marker
Not shown here, see Example1 -
plot-2: [response analysis] Scatter chart
Not shown here, see Example1 -
plot-3: [response analysis] Four-quadrant chart
Not shown here, see Example1 -
plot-4: [response analysis] ROC curve
Not shown here, see Example1 -
plot-5: [survival analysis] survival plot of single marker
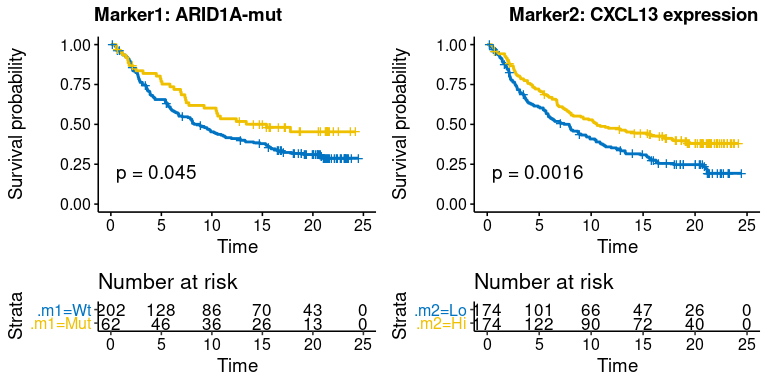
- survival.plot$km.m1m1: KMplot of marker1 and marker2 on two parallel sub-plots
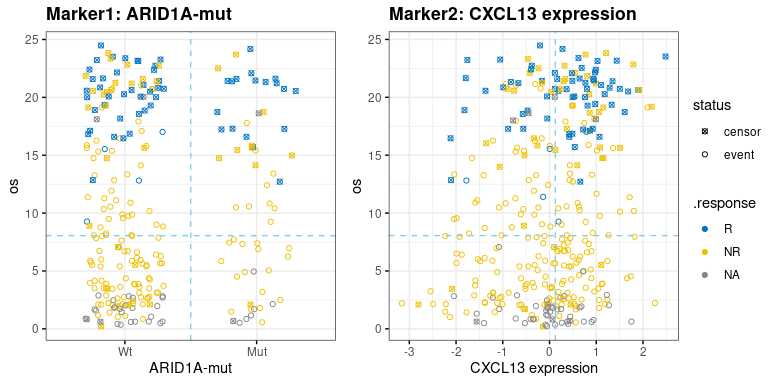
- survival.plot$scatter.m1m2: scatter-plot of survival time(y-axis) and marker1/marker2(x-axis) on two parallel sub-plots, response status(if provided) is shown as color. Nominal survival time is shown and labeled with different shape for the censored data point.
res.pair$survival.plot$km.m1m2res.pair$survival.plot$scatter.m1m2-
plot-6: [survival analysis] survival plot of dual marker
- km.dualmarker: KMplot of dual marker, corresponding to four quadrants in the scatter plot scenario.
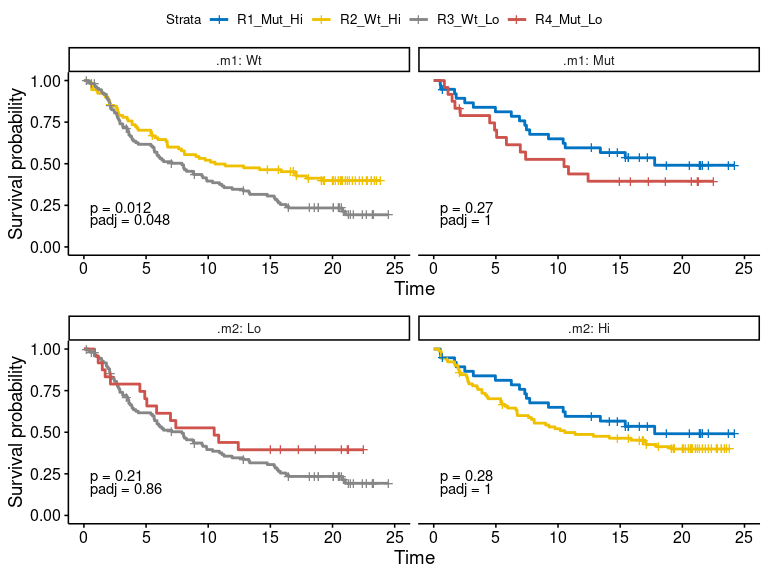
- km.dualmarker.facet: conditional KMplot of dual marker. The conditional KMplot represents the survival curve of marker1 on condition of marker2-level (+/hi or -/lo), and marker2 on condition of marker1 level. It reveals the association between survival and marker1 on the context of marker2 level and vice verse. P-values of log-rank test and adjusted p-values by ‘Bonferroni’ method are shown on each sub-plot.
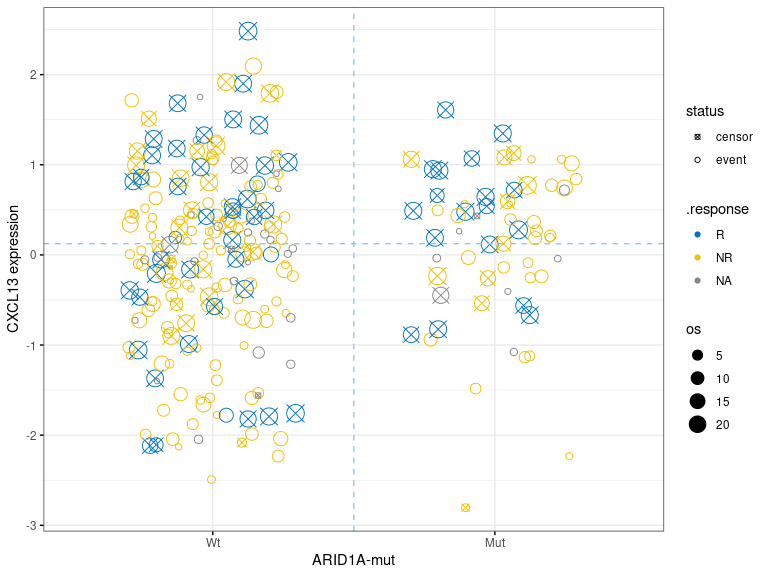
- scatter.dualmarker: scatter-plot of marker1 and marker2 with survival time shown as the size of dot and response (if provided) as color. Nominal survival time is shown and labeled with different shape for the censored data point.
res.pair$survival.plot$km.dualmarkerres.pair$survival.plot$km.dualmarker.facetres.pair$survival.plot$scatter.dualmarker-
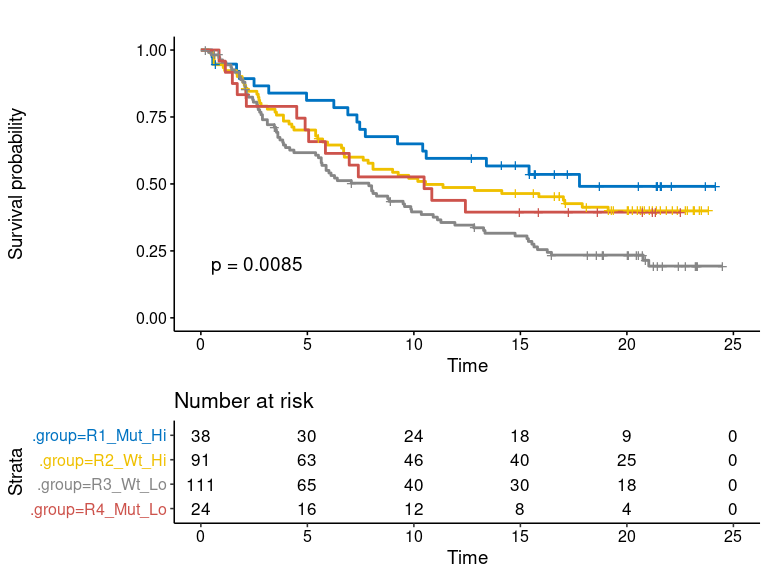
plot-7: [survival analysis] Four-quadrant chart
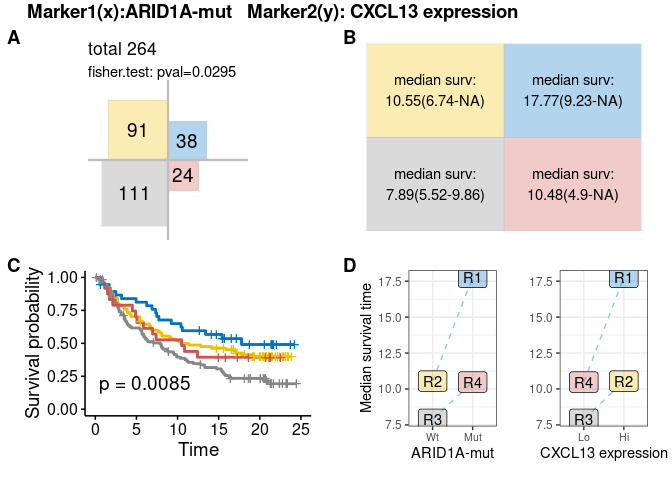
Like response analysis, four-quadrant plots for survival also contains 4 sub-figures.- Area proportion chart: shows the sample size in each quadrant, this chart may be different from counterpart in response analysis owning to different missing samples of survival and response data.
- Statistic matrix: median survival time and confidence interval
- KMplot of four quadrants
- Line chart of median survival time for each quadrant
res.pair$survival.plot$four.quadrant-
stats-1: [response analysis] Four-quadrant statistics
not shown here, see Example1 -
stats-2: [response analysis] Logistic regression results
not shown here, see Example1 -
stats-3: [survival analysis] Four-quadrant statistics
(survival.stats)four.quadrant contains four-quadrant statistics of survival in the following 2 objects:- param contains the note of marker1,marker2, cutoff methods et al.
- stats contains the sample number, median survival and confidence interval in each quadrant, R1-R4
stats.4q <- res.pair$survival.stats$four.quadrant
stats.4q$param
#> # A tibble: 1 x 10
#> time event marker1 marker2 cutpoint.m1 m1.cat.pos m1.cat.neg cutpoint.m2
#> <chr> <chr> <chr> <chr> <lgl> <chr> <chr> <dbl>
#> 1 os cens… mut_AR… gep_CX… NA YES NO 0.124
#> # … with 2 more variables: m2.cat.pos <chr>, m2.cat.neg <chr>
stats.4q$stats
#> # A tibble: 4 x 8
#> .quadrant .m1 .m2 records events median `0.95LCL` `0.95UCL`
#> <fct> <fct> <fct> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 R1 Mut Hi 38 18 17.8 9.23 NA
#> 2 R2 Wt Hi 91 53 10.5 6.74 NA
#> 3 R3 Wt Lo 111 82 7.89 5.52 9.86
#> 4 R4 Mut Lo 24 14 10.5 4.90 NA-
stats-4: [survival analysis] Cox regression result
Like binary outcome (response) analysis, four Cox regression models are built. Model comparison between dual-marker and single-marker models is performed by Likelihood ratio test(LRT) using anova function:- model1: Surv ~ marker1 + covariates, labeled as ‘SM1’ for short
- model2: Surv ~ marker2 + covariates, labeled as ‘SM2’ for short
- model3: Surv ~ marker1 + marker2 + covariates, labeled as ‘DM’ for short, i.e. dual-marker
- model4: Surv ~ marker1 * marker2 + covariates (with interaction term), labeled as ‘DMI’ for short, i.e. dual-marker with interaction
Cox regression models return the following values:
- basic parameters: time, event, m1(marker1), m2(marker2), cut point of m1/m2 if m1/m2 are continuous, positive/negative values if m1/m2 is categorical
- Cox regression parameters: estimate(weight) and p-value(Wald test) of each predictive variable in model1(SM1), model2(SM2), model3(DM), model4(DMI). ‘DMI_m1:m2_estimate’ and ‘DMI_m1:m2_pval’ is estimate and p-value of the interaction term of marker1 and marker2.
- AIC: AIC of SM1, SM2, DM, DMI model
- CPE: concordant probability estimate to evaluate the performance of Cox model using CPE package
- model comparison: p-values of Likelihood Ratio Test(LRT) for SM1-vs-NULL(R ~ 1, no marker), SM2-vs-NULL, SM1-vs-MD, SM2-vs-DM, SM1-vs-DMI, SM2-vs-DMI
dplyr::glimpse(res.pair$survival.stats$cox)
#> Rows: 1
#> Columns: 39
#> $ time <chr> "os"
#> $ even <chr> "censOS"
#> $ m1 <chr> "mut_ARID1A"
#> $ m2 <chr> "gep_CXCL13"
#> $ covariates <chr> ""
#> $ cutpoint_m1 <lgl> NA
#> $ cutpoint_m2 <lgl> NA
#> $ m1_cat_pos <chr> "YES"
#> $ m1_cat_neg <chr> "NO"
#> $ m2_cat_pos <chr> ""
#> $ m2_cat_neg <chr> ""
#> $ SM1_m1_estimate <dbl> -0.3931839
#> $ SM1_m1_p.value <dbl> 0.04572493
#> $ SM2_m2_estimate <dbl> -0.2869317
#> $ SM2_m2_p.value <dbl> 0.0001590714
#> $ DM_m1_estimate <dbl> -0.3134224
#> $ DM_m1_p.value <dbl> 0.1149989
#> $ DM_m2_estimate <dbl> -0.2676217
#> $ DM_m2_p.value <dbl> 0.0004277342
#> $ DMI_m1_estimate <dbl> -0.3045374
#> $ DMI_m1_p.value <dbl> 0.1233496
#> $ DMI_m2_estimate <dbl> -0.2463571
#> $ DMI_m2_p.value <dbl> 0.002164497
#> $ `DMI_m1:m2_estimate` <dbl> -0.1938994
#> $ `DMI_m1:m2_p.value` <dbl> 0.4121259
#> $ SM1_AIC <dbl> 1695.692
#> $ SM2_AIC <dbl> 1686.086
#> $ DM_AIC <dbl> 1685.451
#> $ DMI_AIC <dbl> 1686.794
#> $ SM1_CPE <dbl> 0.535011
#> $ SM2_CPE <dbl> 0.5757988
#> $ DM_CPE <dbl> 0.5834954
#> $ DMI_CPE <dbl> 0.587801
#> $ pval_SM1_vs_NULL <dbl> 0.03779216
#> $ pval_SM2_vs_NULL <dbl> 0.0001907733
#> $ pval_SM1_vs_DM <dbl> 0.0004674887
#> $ pval_SM2_vs_DM <dbl> 0.1044939
#> $ pval_SM1_vs_DMI <dbl> 0.001582535
#> $ pval_SM2_vs_DMI <dbl> 0.1928236Example3: search GEP candidates marker2 to combine with mut_ARID1A for survival analysis
Search among gene expression candidates to combine with ARID1A mutation using dm_searchM2_cox
m2.candidates <- stringr::str_subset(colnames(clin_bmk_IMvigor210),"gep_")
res.m2.cox <- dm_searchM2_cox(
data = clin_bmk_IMvigor210,
# survival
time = "os",
event = "censOS",
# marker1
marker1 = "mut_ARID1A",
m1.binarize = T,
m1.cat.pos = "YES",
m1.cat.neg = "NO",
# marker2
m2.candidates = m2.candidates,
m2.binarize = F, # as continuous variables
p.adjust.method = "BH"
)Glance of the top candidates gene: dm_searchM2_topPlot takes the result of either dm_searchM2_cox or dm_searchM2_logit as input, and returns following figures:
- $m2_effect dot-chart showing top significant marker2s using model comparison dual-vs-marker1
- $interact dot-chart showing top significant marker2s having statistical interaction with marker1
- $m1_m2_effect scatter-plot showing log10-p-value of all marker2s in model comparison of dual-vs-marker1 and dual-vs-marker2
plot.m2.cox <- dm_searchM2_topPlot(res.m2.cox, top.n = 20,
show.padj = F, palette = "jco")- plot-1: marker2 effect
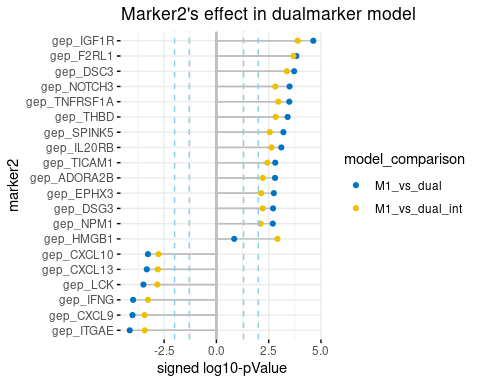
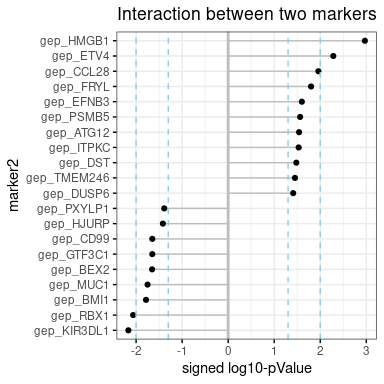
‘m2_effect’ is dot-chart, showing the top significant marker2s, whose introduction to dual-maker model(w/ or w/o interaction) significantly increase the prediction of survival or response. Likelihood ratio test(LRT) is carried out to compare dual-marker model and marker1 single model, the signed log10-pValue is shown on x-axis, and ‘sign’ indicates the effect direction of marker2(single marker) to survival. i.e. genes with negative values on the left are positive survival predictors.
plot.m2.cox$m2_effect- plot-2: marker2’s interaction
‘interaction’ is dot-chart, showing the top significant marker2s, which has statistical interaction with marker1. Signed log10-pValue is shown like ‘m2_effect’
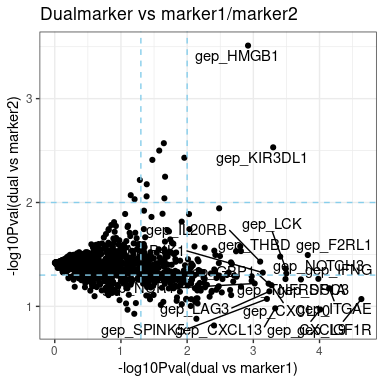
plot.m2.cox$interact- plot-3: m1 and m2 effect
‘m1_m2_effect’ is scatter-plot, showing the -log10-pValue of model comparison, i.e. dual-vs-marker1 and dual-vs-marker2. Dual model that superior to both marker1 and marker2 single marker model is preferred, located top-right on the figure.
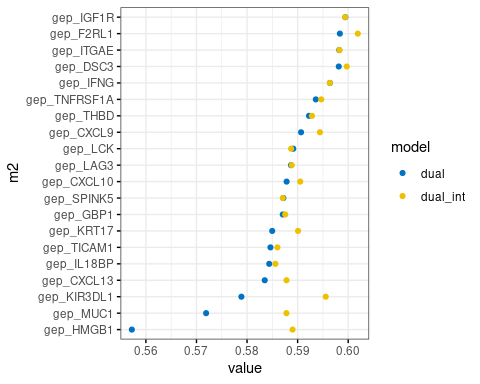
plot.m2.cox$m1_m2_effect- plot-4: CPE ‘CPE’ is concordant probability estimate to evaluate the performance of Cox model using CPE package
plot.m2.cox$CPE- stats: cox result
this shows the same values with res_pair(survival.stats)cox or res_pair(response.stats)logit
dplyr::glimpse(res.m2.cox)
#> Rows: 1,268
#> Columns: 52
#> $ time <chr> "os", "os", "os", "os", "os", "os", "os", "os", …
#> $ even <chr> "censOS", "censOS", "censOS", "censOS", "censOS"…
#> $ m1 <chr> "mut_ARID1A", "mut_ARID1A", "mut_ARID1A", "mut_A…
#> $ m2 <chr> "gep_ADA", "gep_AKT3", "gep_CD24", "gep_BCL2L11"…
#> $ covariates <chr> "", "", "", "", "", "", "", "", "", "", "", "", …
#> $ cutpoint_m1 <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ cutpoint_m2 <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
#> $ m1_cat_pos <chr> "YES", "YES", "YES", "YES", "YES", "YES", "YES",…
#> $ m1_cat_neg <chr> "NO", "NO", "NO", "NO", "NO", "NO", "NO", "NO", …
#> $ m2_cat_pos <chr> "", "", "", "", "", "", "", "", "", "", "", "", …
#> $ m2_cat_neg <chr> "", "", "", "", "", "", "", "", "", "", "", "", …
#> $ SM1_m1_estimate <dbl> -0.3931839, -0.3931839, -0.3931839, -0.3931839, …
#> $ SM1_m1_p.value <dbl> 0.04572493, 0.04572493, 0.04572493, 0.04572493, …
#> $ SM1_m1_p.adj <dbl> 0.04572493, 0.04572493, 0.04572493, 0.04572493, …
#> $ SM2_m2_estimate <dbl> 0.020472278, -0.088602489, -0.040055394, -0.1144…
#> $ SM2_m2_p.value <dbl> 0.7921732808, 0.2316870325, 0.5845798870, 0.1387…
#> $ SM2_m2_p.adj <dbl> 0.90686960, 0.48881723, 0.79339732, 0.37478179, …
#> $ DM_m1_estimate <dbl> -0.3925669, -0.3902232, -0.3992445, -0.3964988, …
#> $ DM_m1_p.value <dbl> 0.04608730, 0.04740445, 0.04273637, 0.04396925, …
#> $ DM_m1_p.adj <dbl> 0.07226057, 0.07226057, 0.07226057, 0.07226057, …
#> $ DM_m2_estimate <dbl> 0.018169157, -0.086696989, -0.048006083, -0.1165…
#> $ DM_m2_p.value <dbl> 0.815536324, 0.243180093, 0.514144401, 0.1321049…
#> $ DM_m2_p.adj <dbl> 0.92247998, 0.51861521, 0.75506892, 0.39294519, …
#> $ DMI_m1_estimate <dbl> -0.4642526, -0.3999853, -0.4101674, -0.3948740, …
#> $ DMI_m1_p.value <dbl> 0.02572344, 0.04411029, 0.03869969, 0.04542806, …
#> $ DMI_m1_p.adj <dbl> 0.08066473, 0.08066473, 0.08066473, 0.08066473, …
#> $ DMI_m2_estimate <dbl> 0.094208982, -0.069846691, -0.092351419, -0.1520…
#> $ DMI_m2_p.value <dbl> 0.284405899, 0.402004262, 0.263768116, 0.0785384…
#> $ DMI_m2_p.adj <dbl> 0.60572337, 0.72716320, 0.58779960, 0.33195570, …
#> $ `DMI_m1:m2_estimate` <dbl> -0.37225167, -0.08245620, 0.22222076, 0.17555057…
#> $ `DMI_m1:m2_p.value` <dbl> 0.05473952, 0.65399403, 0.23758675, 0.35539251, …
#> $ `DMI_m1:m2_p.adj` <dbl> 0.9961319, 0.9961319, 0.9961319, 0.9961319, 0.99…
#> $ SM1_AIC <dbl> 1695.692, 1695.692, 1695.692, 1695.692, 1695.692…
#> $ SM2_AIC <dbl> 1699.937, 1698.583, 1699.707, 1697.803, 1697.684…
#> $ DM_AIC <dbl> 1697.637, 1696.336, 1697.266, 1695.415, 1695.912…
#> $ DMI_AIC <dbl> 1695.951, 1698.136, 1697.843, 1696.573, 1697.739…
#> $ SM1_CPE <dbl> 0.535011, 0.535011, 0.535011, 0.535011, 0.535011…
#> $ SM2_CPE <dbl> 0.5059344, 0.5255417, 0.5106205, 0.5308784, 0.53…
#> $ DM_CPE <dbl> 0.5386091, 0.5507807, 0.5433692, 0.5559563, 0.55…
#> $ DMI_CPE <dbl> 0.5580672, 0.5493288, 0.5512611, 0.5600019, 0.55…
#> $ pval_SM1_vs_NULL <dbl> 0.03779216, 0.03779216, 0.03779216, 0.03779216, …
#> $ padj_SM1_vs_NULL <dbl> 0.03779216, 0.03779216, 0.03779216, 0.03779216, …
#> $ pval_SM2_vs_NULL <dbl> 0.792048437, 0.232822940, 0.584731219, 0.1377636…
#> $ padj_SM2_vs_NULL <dbl> 0.90684524, 0.48877399, 0.79468294, 0.37566516, …
#> $ pval_SM1_vs_DM <dbl> 0.815428198, 0.244342092, 0.514221058, 0.1313353…
#> $ padj_SM1_vs_DM <dbl> 0.92235768, 0.52008877, 0.75466702, 0.39462852, …
#> $ pval_SM2_vs_DM <dbl> 0.03812741, 0.03933856, 0.03508215, 0.03618768, …
#> $ padj_SM2_vs_DM <dbl> 0.06163704, 0.06163704, 0.06163704, 0.06163704, …
#> $ pval_SM1_vs_DMI <dbl> 0.154071909, 0.459326828, 0.396740154, 0.2102864…
#> $ padj_SM1_vs_DMI <dbl> 0.5613885, 0.7924169, 0.7408932, 0.6386168, 0.73…
#> $ pval_SM2_vs_DMI <dbl> 0.01844880, 0.10824010, 0.05327299, 0.07316198, …
#> $ padj_SM2_vs_DMI <dbl> 0.1545506, 0.1545506, 0.1545506, 0.1545506, 0.15…Top 10 dual marker pairs with the highest concordant probability(CPE)
res.m2.cox %>%
dplyr::arrange(desc(DM_CPE)) %>%
dplyr::select(m1, m2, DM_CPE) %>%
head(10) %>%
knitr::kable()| m1 | m2 | DM_CPE |
|---|---|---|
| mut_ARID1A | gep_IGF1R | 0.5994349 |
| mut_ARID1A | gep_F2RL1 | 0.5983506 |
| mut_ARID1A | gep_ITGAE | 0.5982278 |
| mut_ARID1A | gep_DSC3 | 0.5981378 |
| mut_ARID1A | gep_IFNG | 0.5963877 |
| mut_ARID1A | gep_TNFRSF1A | 0.5935954 |
| mut_ARID1A | gep_THBD | 0.5922490 |
| mut_ARID1A | gep_CXCL9 | 0.5906786 |
| mut_ARID1A | gep_LCK | 0.5891381 |
| mut_ARID1A | gep_LAG3 | 0.5886844 |
Example4: evaluation for all combinations
dm_combM_logit and dm_combM_cox will evaluate all combinations of dual-markers.Searching all biomarker pairs is time-consuming for thousands of gene expression biomarker. Here we demonstrate with only 4 biomarkers.
m.candidates <- c("TMB", "gep_CD274","gep_CXCL13", "gepscore_TGFb.19gene", "mut_ARID1A")
res.combM.logit <- dm_combM_logit(
data = clin_bmk_IMvigor210,
response = "binaryResponse",
response.pos = "CR/PR",
response.neg = "SD/PD",
candidates = m.candidates,
m.binarize = F # don't dichotomize marker
)- glimpse of the result
dplyr::glimpse(res.combM.logit)
#> Rows: 10
#> Columns: 61
#> $ response <chr> "binaryResponse", "binaryResponse", "binaryRe…
#> $ response_pos <chr> "CR/PR", "CR/PR", "CR/PR", "CR/PR", "CR/PR", …
#> $ response_neg <chr> "SD/PD", "SD/PD", "SD/PD", "SD/PD", "SD/PD", …
#> $ m1 <chr> "TMB", "TMB", "TMB", "TMB", "gep_CD274", "gep…
#> $ m2 <chr> "gep_CD274", "gep_CXCL13", "gepscore_TGFb.19g…
#> $ covariates <chr> "", "", "", "", "", "", "", "", "", ""
#> $ cutpoint_m1 <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA
#> $ cutpoint_m2 <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA
#> $ m1_cat_pos <chr> "", "", "", "", "", "", "", "", "", ""
#> $ m1_cat_neg <chr> "", "", "", "", "", "", "", "", "", ""
#> $ m2_cat_pos <chr> "", "", "", "", "", "", "", "", "", ""
#> $ m2_cat_neg <chr> "", "", "", "", "", "", "", "", "", ""
#> $ SM1_m1_estimate <dbl> 0.09963989, 0.09963989, 0.09963989, 0.0940112…
#> $ SM1_m1_p.value <dbl> 1.395883e-06, 1.395883e-06, 1.395883e-06, 3.9…
#> $ SM1_m1_p.adj <dbl> 4.652943e-06, 4.652943e-06, 4.652943e-06, 9.8…
#> $ SM2_m2_estimate <dbl> 0.1867642, 0.3217637, -0.1199803, NA, 0.33410…
#> $ SM2_m2_p.value <dbl> 0.20250772, 0.04076421, 0.02516686, NA, 0.020…
#> $ SM2_m2_p.adj <dbl> 0.20250772, 0.04891706, 0.03775028, NA, 0.037…
#> $ DM_m1_estimate <dbl> 0.09848988, 0.09676696, 0.09663392, 0.0941872…
#> $ DM_m1_p.value <dbl> 2.080848e-06, 3.154783e-06, 2.582342e-06, 6.5…
#> $ DM_m1_p.adj <dbl> 1.051594e-05, 1.051594e-05, 1.051594e-05, 1.6…
#> $ DM_m2_estimate <dbl> 0.10743248, 0.23572985, -0.08748326, NA, 0.32…
#> $ DM_m2_p.value <dbl> 0.503579159, 0.162771037, 0.135586952, NA, 0.…
#> $ DM_m2_p.adj <dbl> 0.503579159, 0.195325245, 0.195325245, NA, 0.…
#> $ DMI_m1_estimate <dbl> 0.09700506, 0.09902256, 0.11992599, 0.1137982…
#> $ DMI_m1_p.value <dbl> 6.351301e-06, 2.740248e-05, 1.162081e-06, 6.1…
#> $ DMI_m1_p.adj <dbl> 3.175651e-05, 9.134159e-05, 1.162081e-05, 1.5…
#> $ DMI_m2_estimate <dbl> 0.0444516, 0.2917198, -0.2967303, NA, 0.32866…
#> $ DMI_m2_p.value <dbl> 0.879468167, 0.373035971, 0.003635338, NA, 0.…
#> $ DMI_m2_p.adj <dbl> 0.879468167, 0.447643165, 0.007270677, NA, 0.…
#> $ `DMI_m1:m2_estimate` <dbl> 0.005616161, -0.005515996, 0.018246504, NA, 0…
#> $ `DMI_m1:m2_p.value` <dbl> 0.797807872, 0.841296363, 0.007925505, NA, 0.…
#> $ `DMI_m1:m2_p.adj` <dbl> 0.84129636, 0.84129636, 0.04755303, NA, 0.841…
#> $ SM1_auc <dbl> 0.728, 0.728, 0.728, 0.714, 0.567, 0.567, 0.5…
#> $ SM2_auc <dbl> 0.565, 0.602, 0.591, 0.543, 0.603, 0.596, 0.5…
#> $ DM_auc <dbl> 0.726, 0.738, 0.747, 0.712, 0.602, 0.642, 0.5…
#> $ DMI_auc <dbl> 0.729, 0.738, 0.754, 0.718, 0.601, 0.650, 0.5…
#> $ SM1_AIC <dbl> 239.2481, 239.2481, 239.2481, 234.5140, 321.8…
#> $ SM2_AIC <dbl> 270.8963, 268.1304, 267.3176, 262.5763, 318.3…
#> $ DM_AIC <dbl> 240.8011, 239.2501, 238.9800, 236.5126, 320.3…
#> $ DMI_AIC <dbl> 242.7342, 241.2101, 233.9904, 237.2973, 322.2…
#> $ pval_SM1_vs_NULL <dbl> 7.993656e-09, 7.993656e-09, 7.993656e-09, 4.7…
#> $ padj_SM1_vs_NULL <dbl> 2.664552e-08, 2.664552e-08, 2.664552e-08, 1.1…
#> $ pval_SM2_vs_NULL <dbl> 0.20191142, 0.03605771, 0.02249407, 0.1841746…
#> $ padj_SM2_vs_NULL <dbl> 0.20191142, 0.07211542, 0.05623518, 0.2019114…
#> $ pval_SM1_vs_DM <dbl> 0.5037288824, 0.1575035521, 0.1320594393, 0.9…
#> $ padj_SM1_vs_DM <dbl> 0.559698758, 0.313680337, 0.313680337, 0.9697…
#> $ pval_SM2_vs_DM <dbl> 1.467972e-08, 2.744417e-08, 3.630161e-08, 1.1…
#> $ padj_SM2_vs_DM <dbl> 1.210054e-07, 1.210054e-07, 1.210054e-07, 2.9…
#> $ pval_SM1_vs_DMI <dbl> 0.773389957, 0.360955160, 0.009765815, 0.5442…
#> $ padj_SM1_vs_DMI <dbl> 0.77338996, 0.60472781, 0.03255272, 0.6047278…
#> $ pval_SM2_vs_DMI <dbl> 1.037739e-07, 1.930837e-07, 7.843230e-09, 4.3…
#> $ padj_SM2_vs_DMI <dbl> 5.188693e-07, 6.436125e-07, 7.843230e-08, 1.0…
#> $ SM2_m2YES_estimate <dbl> NA, NA, NA, 0.4571665, NA, NA, 0.4443805, NA,…
#> $ SM2_m2YES_p.value <dbl> NA, NA, NA, 0.1789181, NA, NA, 0.1896272, NA,…
#> $ DM_m2YES_estimate <dbl> NA, NA, NA, -0.01465024, NA, NA, 0.43898759, …
#> $ DM_m2YES_p.value <dbl> NA, NA, NA, 0.9697412, NA, NA, 0.1970138, NA,…
#> $ DMI_m2YES_estimate <dbl> NA, NA, NA, 0.5892055, NA, NA, 0.4397188, NA,…
#> $ DMI_m2YES_p.value <dbl> NA, NA, NA, 0.3682456, NA, NA, 0.1967279, NA,…
#> $ `DMI_m1:m2YES_estimate` <dbl> NA, NA, NA, -0.04543200, NA, NA, -0.01333611,…
#> $ `DMI_m1:m2YES_p.value` <dbl> NA, NA, NA, 0.2625779, NA, NA, 0.9671296, NA,…Users can filter and get interesting dual-marker pairs using plenty of
statistics and model performance metrics, for example, AUC for response
analysis (logistic regression), concordant probability CPE for survival
analysis (Cox regression), p-value of dual-vs-single model comparison
and statistical interaction of two markers.
Top 2 dual-marker pairs with the highest AUC
res.combM.logit %>%
arrange(desc(DM_auc)) %>%
dplyr::select(m1, m2, DM_auc) %>%
head(2) %>%
knitr::kable()| m1 | m2 | DM_auc |
|---|---|---|
| TMB | gepscore_TGFb.19gene | 0.747 |
| TMB | gep_CXCL13 | 0.738 |