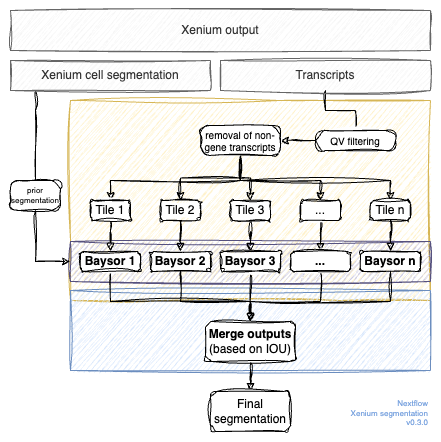
A nextflow pipeline for segmentation of 10x Xenium runs. The pipeline consists of the following parts:
- New step in the pipeline to run xenium ranger with the new segmentation to allow of exploration of the data using Xenium Explorer (This automatically removes cells with less than 3 transcripts)
The pipeline has been adjusted to use the files generated from Xenium running software version 3. The main changes are:
- The pipeline no longer runs the custom nuclear segmentation, as the multimodal segmentation kit yields good results.
- The results from the multimodal segmentation are now passed onto Baysor as a prior segmentation.
- The scripts have been adapted to use the parquet files instead of csv files.
- Filter the transcripts based on OV values, remove non-gene transcripts and create smaller tiles of the dataset. For more information see tile-xenium.
- Run Baysor on all tiles. This step uses Baysor wrapped in a docker image.
- Merge the segmentation results to get one final segmentation. This program merges cells based on an IOU threshold.
The pipeline requires a working installation of nextflow. Further, it uses singularity/apptainer to run the scripts in reproducible environments. Run the pipeline with:
nextflow run maximilian-heeg/xenium-segmentation -r v0.3.1 --xenium_path PATH_TO_XENIUM_OUTOUT
Many parameters can be manually set (and might need to be adjusted for best reuslts.)
/*********************************
Input and Output.
*********************************/
// The folder pointing towards the xenium results
xenium_path = null
// Path to where the final csv should be saved.
outdir = 'results'
/*********************************
Tile creation
*********************************/
// The width of a tile
tile.width = 4000
// The height of a tile
tile.height = 4000
// The overlap between titles. The next tile with start at x + width - overlap,
// or y + height - overlap
tile.overlap = 200
// The minimum Q-Score to pass filtering.
tile.qv = 20
// The minimum number of transcripts per tile. This is important
// because baysor cannot work, if there are too few transcripts.
// If the minimal number is not met in a tile, it will be expanded by `overlap`
// in all fofur directions until the criterion is met.
tile.minimal_transcripts = 10000
/*********************************
Baysor
*********************************/
// Minimal number of molecules for a cell to be considered as
// real. It's an important parameter, as it's used to infer
// several other parameters. Overrides the config value.
// If set to -1, the pipeline will calcuate the median transcripts per cell based
// on the 10x segmentation and use that value as min_molecules_per_cell
baysor.min_molecules_per_cell = -1
// If min_molecules_per_cell should be estimated, this values defines the fraction of
// the median transcripts per cell that will be set as min_molecules_per_cell
baysor.min_molecules_per_cell_fraction = 0.75
// Minimal number of molecules in a segmented region, required for this region to be
// considered as a possible cell.
// 0: min_molecules_per_cell / 4 (Baysor default)
// string ended with '%' to set it relative to min_molecules_per_cell
baysor.min_molecules_per_segment = '50%'
// Standard deviation of scale across cells. Can be either
// number, which means absolute value of the std, or string
// ended with '%' to set it relative to scale (default: "25%")
// Negative values mean it must be estimated from `min_molecules_per_cell`
baysor.scale = -1.0
// Standard deviation of scale across cells. Can be either number, which
// means absolute value of the std, or string ended with "%" to set it
// relative to scale. Default: "25%"
baysor.scale_std = "25%"
// Number of clusters to use for cell type segmentation. Default: 4
baysor.n_clusters = 4
// Confidence of the prior_segmentation results.
// Value in [0; 1]. If you want the final segmentation not
// contradicting to prior_segmentation, set it to 1.
// Otherwise, if you assume errors in prior_segmentation,
// values in [0.2-0.7] allow flexibility for the algorithm.
baysor.prior_segmentation_confidence = 0.5
// Comma-separated list of nuclei-specific genes. If provided,
// `cyto-genes` has to be set, as well.
nuclei_genes = ""
// Comma-separated list of cytoplasm-specific genes. If provided,
// `nuclei-genes` has to be set, as well.
cyto_genes = ""
// Comma-separated list of genes or regular expressions to ignore
// during segmentation. Example: 'Blank*,MALAT1'
baysor.exclude_genes = ""
// new-component-weight is proportional to the probability of
// generating a new cell for a molecule, instead of assigning
// it to one of the existing cells. More precisely, the probability
// to assign a molecule to a particular cell linearly depends on
// the number of molecules, already assigned to this cell. And this
// parameter is used as the number of molecules for a cell, which is
// just generated for this new molecule. The algorithm is robust to
// small changes in this parameter. And normally values in the range
// of 0.1-0.9 should work fine. Smaller values would lead to slower
// convergence of the algorithm, while larger values force the
// emergence of a large number of small cells on each iteration,
// which can produce noise in the result. In general, the default
// value should work well.
baysor.new_component_weight = 0.2
/*********************************
Merging
*********************************/
// Threshold for stitching. If the IOU for two cells is greater than
// the threshold, they will be merged
merge.iou_threshold = 0.2
/*********************************
Xenium Ranger
*********************************/
// alpha value for alphashape calculation.
// this is used to calcuate the boundaries of the cells based
// on the transcripts
xeniumranger.alpha = 0.05
These parameters can either be changed by adding the parameters to the command (--baysor.min_molecules_per_cell 60) or by creating a nextflow.config file within the working directory.
// Change the min_molecules_per_cell for Baysor
params {
baysor.min_molecules_per_cell = 60
}
In the Nextflow framework architecture, the executor is the component that determines the system where a pipeline process is run and supervises its execution.
The executor provides an abstraction between the pipeline processes and the underlying execution system. This allows you to write the pipeline functional logic independently from the actual processing platform.
In other words, you can write your pipeline script once and have it running on your computer, a cluster resource manager, or the cloud — simply change the executor definition in the Nextflow configuration file.
Again, you can set an executor by modifying the nextflow.config.
// Use the PBS scheduler
process {
executor = 'pbs'
queue = 'home-yeo'
}
Since Nextflow version 23.07.0, nextflow no longer mounts the home directory when launching a Singularity container. This can cause some errors with the cache directories in python scripts, but can be fixed by setting export NXF_SINGULARITY_HOME_MOUNT=true.