V. Vedder and T. Reinberger et al.
"pyHeart4Fish: Chamber-specific heart phenotype quantification of zebrafish in high-content screens"
Front. Cell Dev. Biol. Sec. Molecular and Cellular Pathology , Volume 11 - 2023 | doi: 10.3389/fcell.2023.1143852
- How to install pyHeart4Fish
- Tutorial - How to run pyHeart4Fish
- Troubleshooting and FAQ
- Licensing
The pyHeart4Fish has been developed and tested in Windows 10 and 11!
If not done yet download
and install python >3.7 on your computer, please! Check ADD TO PATH when asked.
- Download pyHeart4Fish_python folder from GitHub/ToReinberger/pyHeart4Fish
- Move the complete folder from ./downloads to a desired storage place
(e.g., C:\Users<user_name>\Desktop\pyHeart4Fish_python) - Open command line in Windows (e.g., by typing
cmdin Windows search field) - Move to the pyHeart4Fish folder by typing
cd C:\Users\<YOUR_USERNAME>\Desktop\pyHeart4Fish_scripts - Install required packages by typing
pip install -r requirements.txt
orpip install -r requirements_for_installation.txt
- Download
install_pyHeart4Fish.pyfrom GitHub/ToReinberger/pyHeart4Fish - Start
install_pyHeart4Fish.py> double click OR right click > open with Python - Follow instructions
- aicsimageio==4.7.0
- czifile==2019.7.2
- aicspylibczi==3.0.5
- matplotlib>=3.5.1
- numpy==1.21.2
- opencv_python==4.5.5.62
- pandas>=1.3.4
- Pillow>=9.2.0
- scipy==1.8.0
- openpyxl==3.0.10
11-05-2023
- CziReader from AICSImageIO has been fully replaced czifile package > now all image types can be loaded correctly
- pxiel values of images are now normalied [0 - 255] using cv2.normalize method
- .feather output per fish was replaced with .csv, as .feather cannot store single line outputs
- If a Desktop shortcut has been created, double-click the
pyHeart4Fishicon (Installation option 2) - OR right click on
pyHeart4Fish_scripts/heart_beat_GUI.py> open with python (Installation option 1) - OR in Windows console
python <YOUR_STORAGE_PATH>/pyHeart4Fish_scripts/heart_beat_GUI.py(Installation option 2) - OR start
heart_beat_GUI.pyin a Python IDE of your choice (e.g., Pycharm, KITE, Notepad++) - OR double-click on
pyHeart4Fish_exe/heart_beat_GUI.exe(Installation option 3)
This Python script opens the main window of pyHeart4Fish (see Step 2)
Please check experimental requirements before using pyHeart4Fish:
- Only one zebrafish heart should be imaged at once!
- The optimal acquisition time is between 15 s and 30 s!
- Make sure that the acquisition time is a bit longer as desired (e.g., 22 s)
- Use a frame rate higher than 15 f/s to increase analysis quality for arrhythmias
Accepted file formats:
- .avi
- .czi (ZEISS file format)
- .mp4
- .tif-files, .png-files or .jpg-files as a series of frames stored in one folder per fish
- Config-file as json-file
- Raw data of heartbeats as numpy array
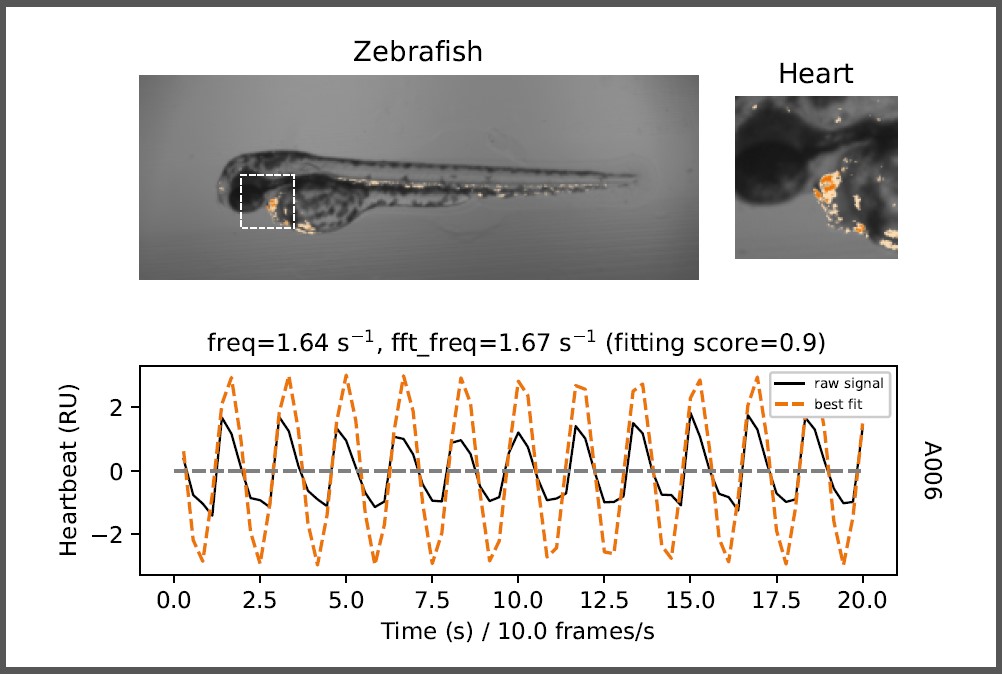
- Atrium and ventricle curves and fitted sine functions as plot
- Processed heart images as GIF-file (10 iterations)
- Excel sheet for each fish in subfolder with all parameters
- Combined Excel sheet for all zebrafish analyzed
Test data can be found on OneDrive.
Please unzip data sets before analysis!
- fish_1_2_avi_files.zip and avi_files_Results_example
- Frames per seconds = 29 and cut movie at = 20 sec
- fish_1_2_czi_files.zip and czi-files_Results_example
- Frames per seconds = 9.5 and cut movie at = 20 sec
- fish_1_2_tif_files and tif_files_Results_example
- Frames per seconds = 29 and cut movie at = 20 sec
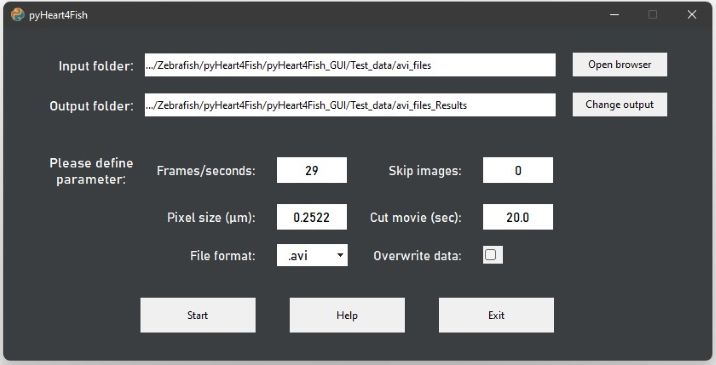
| Input folder: | > Contains all movie-files/ images in sub-folders for one project/ experiment > Choose file folder and click Open Folder |
| Output folder: |
> Is automatically created : Input + "_Results" > Or click Change output to define a desired output folder > Contains all configuration files, raw data, plots and excel sheets (see above) |
| Frames per second: | > Should be > 15 for optimal results for optimal results, but 10 f/s can be sufficient |
| Skip images: | > Default 0, (0 - 10 possible) > 1 = Every second frame is skipped > Might accelerate the analysis as the number of images is cut in half > Use only if frame rate is high enough! |
| Pixel size: | > Please check the size of a pixel at your microscope; in your metadata; or use a scale and ImageJ/Fiji to determine your pixel size [µm] > A wrong pixel size will give wrong heart size etc. > Relative values will still apply |
| Cut movie (sec): | > The length of the movie should be at least 10 s > Ensures that all movies have the same length |
| File format: | > See Input data types |
| Overwrite data: | > If data needs to be re-analyzed > If unselected all analyzed hearts will be skipped in the project folder |
| Acquisition mode: | > Fluorescence (chamber-specific): Fluorescent heart is required and 10x magnification is desired > Bright field (only heartbeat): The script is optimized for bright field movies /series of frames of whole zebrafish in sagittal position (eyes included) |
By clicking Start, the method StartConfigs.run_program is executed which iteratively
executes heart_beat_GUI_only_one_fish_multiprocessing.py
or heart_beat_GUI_only_one_fish_multiprocessing.py
for each zebrafish and combines all Excel sheets once all zebrafish have been analyzed.
In the bright field acquisition mode, all zebrafish will be analyzed automatically.
This will take a while, e.g., 20 to 30 min for 40 zebrafish depending on the computer performance.
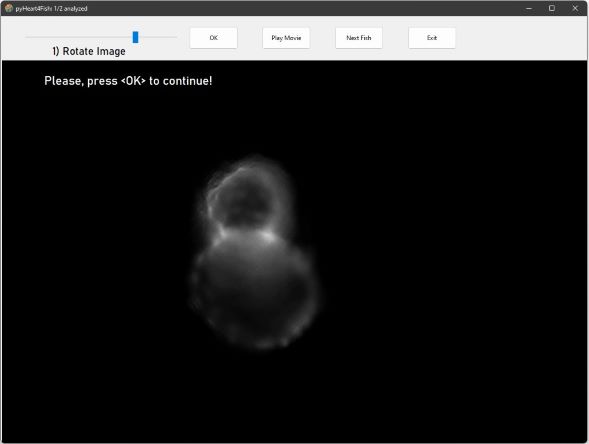
Rotate the heart for chamber-specific fluorescent acquisition mode using the slider to position ventricle
at the top and atrium at the bottom. Click OK to continue.
You can orientate the fish in another direction.
However, the tool will later automatically re-rotate the video for analysis!
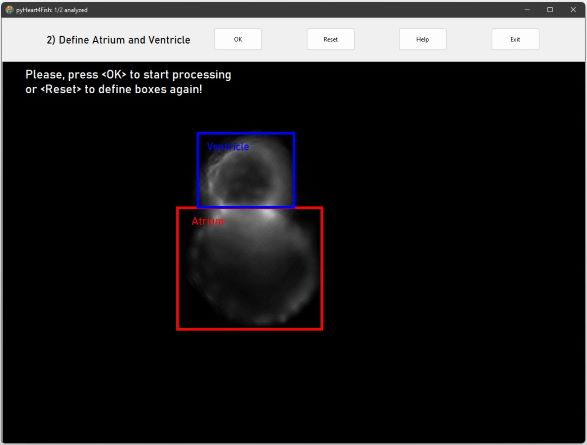
To distinguish between background and heart define 1) atrium and 2) ventricle area by Drag-and-Draw.
Click OK to start the analysis of all frames / images.
Best results yields a rough selection around respective heart chambers (see image below).
Make sure that the border encloses all parts of the atrium!
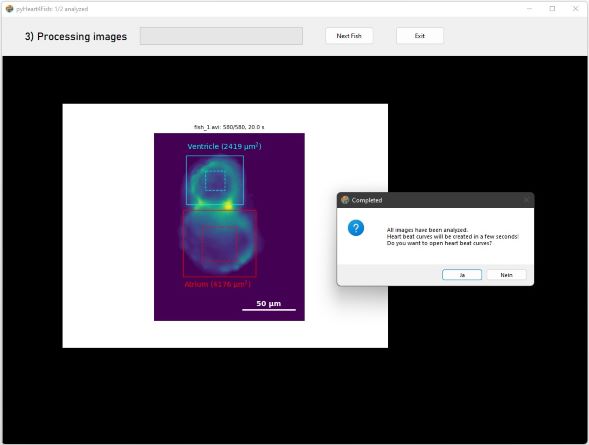
The first processed image is shown after the complete analysis.
Press Yes to show heartbeat curves.
The number of analyzed fish hearts is shown in the top left corner (here: 1/2 analyzed)
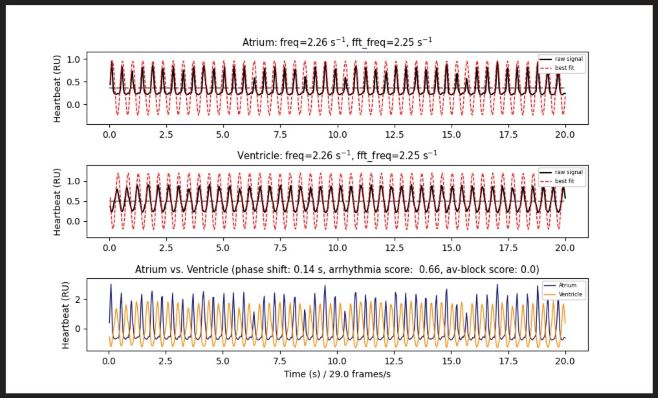
| freq: | > Frequency / rate of heartbeats derived from fitted sine function (red dotted line) |
| fft_freq: |
> Frequency / rate of heartbeats derived from fast fourier transformation (FFT) |
| phase shift: | > Shift between atrium and ventricle sine function > A very small or very high phase shift can be a sign of arrhythmia / AV-block but 10 f/s can be sufficient |
| arrhythmia score: | > The lower this value, the more regular the heartbeat. > A value >0.7 is a sign of arrhythmia |
| av-block score: | > The absolute difference of all frequencies (freq and fft_freq) between atrium and ventricle > A value >0.5 is a sign for a conduction defect or atrial/ ventricle tachycardia or bradycardia |
Once all fish hearts have been analyzed you can choose to open the summary Excel sheet for all fish
Output parameters:
| Project_name | > Name of the project folder |
| Condition |
> File name of of movie / image folder |
| Number_images | > Number of images/ frames analyzed |
| Heart_size (µm^2) | > Size of the heart (atrium + ventricle) |
| x_distance (µm) | > Maximal horizontal distance |
| y_distance (µm) | > Maximal vertical distance |
| Round_shape | > Derived from x_distance-y_distance ratio (1 = round, 0 = stretched) |
| Atrium/ ventricle maximal dilated or maximal contracted chamber area | > Minimal area = contraction; maximal area = dilatation |
| Atrium/ ventricle relative contractility (%) | > Maximal difference in area normalized to maximal chamber area |
| Atrium/ ventricle ejection fraction (µm^3) | > Absolute change in volume (cylinder approximation) |
| Atrium/ ventricle ejection fraction (µm^3) | > Absolute change in volume (cylinder approximation) |
| Atrium/ ventricle fit_score | > Correlation coefficient between raw data and fitted curve (red line) |
| Atrium/ ventricle Auto_Corr | > Auto-Correlation coefficient for raw data > The higher the value, the more regular the heartbeat |
| Freq, Freq_fft, phase_shift, arrhythmia score, av_block_score | > see above |
- If
install_pyHeart4Fish.pydoes not run to the end or the desktop icon is missing,
install the packagepyshortcutsfirst by typingpip install pyshortcutsin Windows console
and run againinstall_pyHeart4Fish.py - You might also install all packages in
requirements.txtseparately from (see GitHub/ToReinberger/pyHeart4Fish
by typingpip install <NAME_OF_PACKAGE>in Windows console
- Make sure that the background fluorescence is as low as possible
- In step 3 and 4, make sure you correctly select the atrium (in most case less bright part of the fish),
otherwise the background threshold is set incorrectly - If the heart hasn't been identified correctly, please try to re-define atrium and ventricle area, e.g., by enlarging the area
- If progress bar is on hold, please click
Next Fishas an error has occurred
Please contact tobias.reinberger@uni-luebeck.de to report any issues
BSD 2-Clause License
Copyright (c) 2022, Tobias Reinberger and Viviana Vedder. All rights reserved.
Cite as V. Vedder and T. Reinberger et al., pyHeart4Fish: Chamber-specific heart phenotype quantification of zebrafish in high-content screens, Front. Cell Dev. Biol. Sec. Molecular and Cellular Pathology , Volume 11 - 2023 | doi: 10.3389/fcell.2023.1143852
Redistribution and use in source and binary forms, with or without modification, are permitted provided that the following conditions are met:
- Redistributions of source code must retain the above copyright notice, this list of conditions and the following disclaimer.
- Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the following disclaimer in the documentation and/or other materials provided with the distribution.
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.