This project contains a feature selection method by genetic algorithm for cheminformatics applications (also applicable to other datasets). The method only needs SMILES for cheminformatics applications. Using Mordred package [1], it generates around 200 features. Genetic algorithm selects the most effective features among them. The repository provides example applications on the following 4 datasets (2 molecular, 2 toy datasets).
Classification:
- BBBP [2] (Blood-Brain Barrier Penetration dataset)
- Iris (Iris plants dataset)
Regression:
- AqSolDB [3] (Aqueous Solubility dataset)
- Boston (Boston house prices dataset)
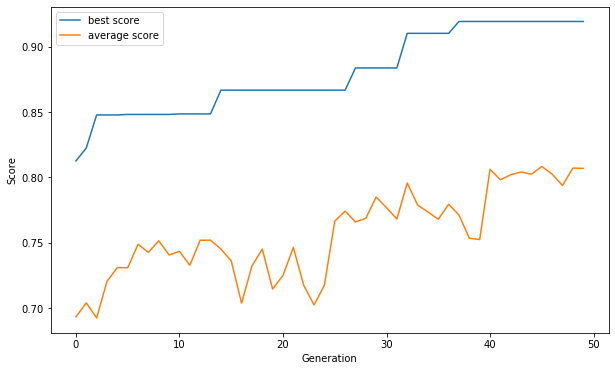
Genetic algorithms are heuristic methods of optimization, inspired by evolution. Basically, it aims to create more successful individuals by transferring the best genes to new generations (see image below). In this application, the genetic algorithm is implemented to select the features that best represent the target.
Each feature is represented as a gene [0 or 1] and the set of features as a chromosome. The score (fitness) of each chromosome is determined by the cross-validation result of training. We used LogisticRegresssion() for classification tasks and LinearRegression() for regression tasks while evaluating the fitness. (more advanced methods can be used for more accurate fitness, but the time cost must be considered.)
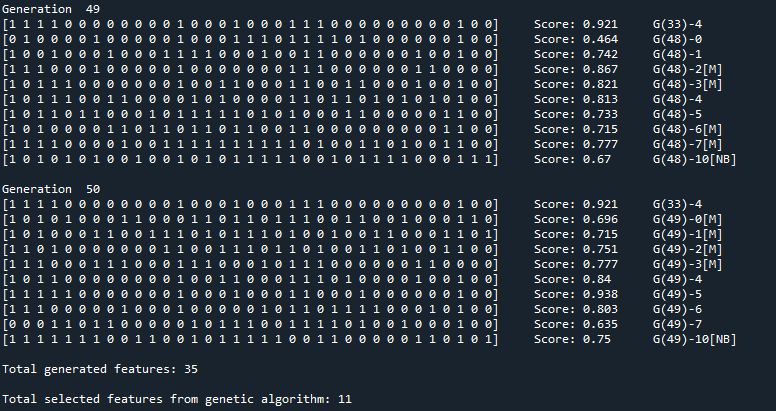
The output can be controlled via "verbose" parameter. While verbose=1, the result of each generation is printed.
- G(X) = Generation number
- [M] = Mutant
- [NB] = New blood
- rdkit==2020.03.2
- mordred==1.2.0
- scikit-learn==0.23.1
- pandas==1.0.3
[1] Moriwaki, Hirotomo, et al. "Mordred: a molecular descriptor calculator." Journal of cheminformatics 10.1 (2018): 4.
[2] Martins, Ines Filipa, et al. "A Bayesian approach to in silico blood-brain barrier penetration modeling." Journal of chemical information and modeling 52.6 (2012): 1686-1697.
[3] Sorkun, Murat Cihan, Abhishek Khetan, and Süleyman Er. "AqSolDB, a curated reference set of aqueous solubility and 2D descriptors for a diverse set of compounds." Scientific data 6.1 (2019): 1-8.