Developed by Tashrif Billah and Sylvain Bouix, Brigham and Women's Hospital (Harvard Medical School)
-
Running instruction is available at TUTORIAL.md.
-
This pipeline is also available as Docker and Singularity containers. See pnlpipe-containers for details.
- Luigi pipeline
- Citation
- Installation
- Tests
- How Luigi works
- Running luigi-pnlpipe
- PNL Luigi server
- Example commands
- Advanced
- Workflows
- Parameters
- Caveat/Issues
Table of Contents created by gh-md-toc
Luigi is a Python module for building complex pipeline of jobs. Psychiatry Neuroimaging Laboratory (PNL) has developed and tested many software modules for MRI processing over years. The individual modules are gracefully joined together using Luigi. With the release of luigi-pnlpipe, researchers should be able to perform MRI processing more elegantly.
If this pipeline is useful in your research, please cite as below:
Billah, Tashrif; Bouix, Sylvain, A Luigi workflow joining individual modules of an MRI processing pipeline, https://github.com/pnlbwh/luigi-pnlpipe, 2020, DOI: 10.5281/zenodo.3666802
The pipeline depends on external software. Please follow this instruction to install those software. Then, you can clone luigi-pnlpipe as follows:
git clone https://github.com/pnlbwh/luigi-pnlpipe.git
NOTE luigi-pnlpipe and pnlNipype must be cloned in the same directory. It is essential because
the former leverages upon soft-linked (ln -s) scripts from the latter.
The pipeline is built upon https://github.com/pnlbwh/pnlNipype individual modules. Documentation about individual pnlNipype modules can be found in pnlNipype/TUTORIAL.md. Finally, see here for instructions about running the pipeline.
A client (PNL external collaborator) can use the official luigi package installed in pnlpipe3 conda environment.
However, a server should install Tashrif's development for /history/by_task_id/ URL to function:
pip install git+https://github.com/tashrifbillah/luigi.git@89c9aa750de8ae2badabe435d98c02e44a1aa8b4
luigi-pnlpipe itself will not fail without Tashrif's development on the server side. That means, you can
also use the official luigi package on the server side. But you will not be able to redirect to
/history/by_task_id/ URLs generated in *.log.html provenance files. Notably, the provenance files
are generated on the client side by _provenance.py when luigi-pipeline is run.
- Clone it
git clone https://github.com/pnlbwh/CNN-Diffusion-MRIBrain-Segmentation.git
- Download models
https://github.com/pnlbwh/CNN-Diffusion-MRIBrain-Segmentation#4-download-models
- Add to PATH
export PATH=/path/to/CNN-Diffusion-MRIBrain-Segmentation/pipeline/:$PATH
Firstly, this requirement is optional. Secondly, it is only required if you would like to run Wma800 task
from our luigi-pnlpipe. The latest whitematteranalysis software comes with a
script that performs whole-brain tractography parcellation.
That script alone imposes a number of dependecies on luigi-pipeline:
- Slicer
Get the stable Slicer from here, unzip it, launch ./Slicer, and finally click on the E like icon on top-right
to install SlicerDMRI extension module.
- Xvfb
The above script needs to launch Slicer under the hood. For that to happen, either run that script within a virtual desktop (NoMachine) or
install xorg-x11-server-Xvfb.
- An atlas
After you have installed whitematteranalysis through our conda recipe, download the required atlas as follows:
wm_download_anatomically_curated_atlas.py -atlas ORG-800FC-100HCP ./
The pipeline is tested nightly via an isolated Docker container. Testing instruction is available here. Tests are designed to be fairly straight forward. However, you may need some Docker and git knowledge to be able to run the tests.
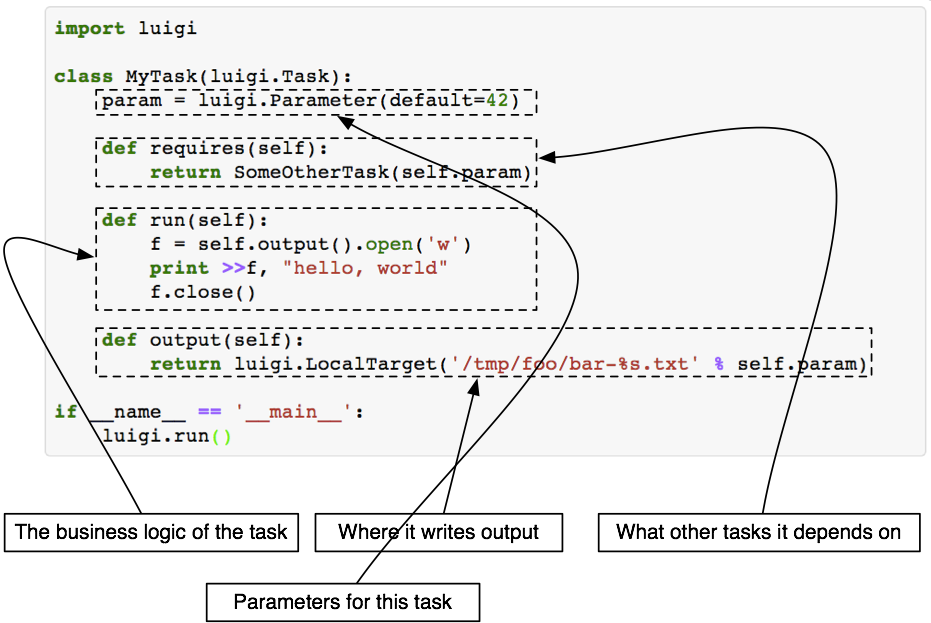
A Luigi task consists of four building blocks:
i) Parameters
ii) requires()
iii) run()
iv) output()
Each individual PNL module is wrapped with a Luigi task. Once a task is scheduled i.e. commanded to execute, Luigi first looks for the output() in local directory.
If the output() does not exist, it starts running the task. Again, it first runs the prerequisite (requires()) of the task.
Then the task itself (run()).
The merit of Luigi task- it won't repeat a task if the output() already exists. On the other hand, the demerit-
if output of a prior requires() task is missing, Luigi will not go back to checking that provided the output() of the current task exists.
So, to force re-run all tasks, you would have to manually delete all intermediate outputs.
The most notable feature of Luigi tool is, it comes with a web visualizer where you can monitor progress of the scheduled tasks.
Another useful feature is multi-worker task scheduling where you can specify number of Luigi workers and each worker can run tasks applying parellel processing. It is like you open multiple terminals on your machine and use multi-processing independently in each of the terminals. Above all, progress of task is reported to the central scheduler that you can view on your web browser.
luigi-pnlpipe makes use of Brain Imaging Data Structure (BIDS). Before running the luigi-pnlpipe, you should organize your data according to BIDS convention. For example, the following is a minimal BIDS organization:
tree ~/Downloads/INTRuST_BIDS
caselist.txt
derivatives
sub-003GNX007
├── anat
│ ├── sub-003GNX007_T1w.nii.gz
│ └── sub-003GNX007_T2w.nii.gz
└── dwi
├── sub-003GNX007_dwi.bval
├── sub-003GNX007_dwi.bvec
└── sub-003GNX007_dwi.nii.gz
sub-003GNX012
├── anat
│ ├── sub-003GNX012_T1w.nii.gz
│ └── sub-003GNX012_T2w.nii.gz
└── dwi
├── sub-003GNX012_dwi.bval
├── sub-003GNX012_dwi.bvec
└── sub-003GNX012_dwi.nii.gz
sub-003GNX021
├── anat
│ ├── sub-003GNX021_T1w.nii.gz
│ └── sub-003GNX021_T2w.nii.gz
└── dwi
├── sub-003GNX021_dwi.bval
├── sub-003GNX021_dwi.bvec
└── sub-003GNX021_dwi.nii.gzOnce data is organized as above, you can define the following arguments:
--bids-data-dir ~/Downloads/INTRuST_BIDS
-c ~/Downloads/INTRuST_BIDS/caselist.txt
--t1-template sub-*/anat/*_T1w.nii.gz
--t2-template sub-*/anat/*_T2w.nii.gz
--dwi-template sub-*/dwi/*_dwi.nii.gz
When luigi-pnlpipe globs bids-data-dir/sub-*/anat/*_T1w.nii.gz replacing * in sub-*
with each line from caselist.txt, it will find the input data.
Finally, output will go to derivatives/pnlpipe directory by default, again following BIDS convention.
Please follow the installation instruction as noted here. Upon successful installation of all software modules,
source .bashrc to set up your terminal for running luigid.
In the packaged version, tasks are configured to use PNL hosted public luigid server:
default-scheduler-url = https://${LUIGI_USERNAME}:${LUIGI_PASSWORD}@pnlservers.bwh.harvard.edu/luigi/Please email tbillah@bwh.harvard.edu to obtain the username and password. They must be defined as environment variables before running luigi-pnlpipe tasks:
export LUIGI_USERNAME=hello
export LUIGI_PASSWORD=world
We strongly recommend using PNL hosted public luigid server for running luigi-pnlpipe tasks inside Docker and Singularity containers.
Detailed instruction is given at pnlpipe-containers/README.md#luigi-tasks.
Instead, you can set up your own luigid server. A useful configuration for luigid server is given in luigi-pnlpipe/luigi.cfg.
For luigid to find that configuration, you should change directory and initiate server as follows:
cd ~/luigi-pnlpipe/
luigid --background --logdir /tmp/luigi-server.log
Upon launching luigid server as above, you must edit luigi.cfg so tasks know to use your local server:
# for individual machine
default-scheduler-url = http://localhost:8082/
...
...
# PNL hosted public server
# default-scheduler-url = https://${LUIGI_USERNAME}:${LUIGI_PASSWORD}@pnlservers.bwh.harvard.edu/luigi/You may also edit luigi.cfg as you see fit.
For running all workflows of luigi-pnlpipe from one place, we have developed the following
handy script. luigi-pnlpipe/exec/ExecuteTask is a soft link for luigi-pnlpipe/workflows/ExecuteTask.py.
You can use one or the other as you like. At this point, you should be able to see the help message. Please take a
moment to familiarize yourself about its functionality.
luigi-pnlpipe/exec/ExecuteTask -h
usage: ExecuteTask.py [-h] --bids-data-dir BIDS_DATA_DIR -c C -s S
[--dwi-template DWI_TEMPLATE]
[--t1-template T1_TEMPLATE] [--t2-template T2_TEMPLATE]
--task
{StructMask,Freesurfer,CnnMask,PnlEddy,PnlEddyEpi,FslEddy,FslEddyEpi,TopupEddy,PnlEddyUkf,Fs2Dwi,Wmql,Wmqlqc}
[--num-workers NUM_WORKERS]
[--derivatives-name DERIVATIVES_NAME]
pnlpipe glued together using Luigi, optional parameters can be set by
environment variable LUIGI_CONFIG_PATH, see luigi-pnlpipe/scripts/params/*.cfg
as example
optional arguments:
-h, --help show this help message and exit
--bids-data-dir BIDS_DATA_DIR
/path/to/bids/data/directory
-c C a single case ID or a .txt file where each line is a
case ID
-s S a single session ID or a .txt file where each line is
a session ID (default: 1)
--dwi-template DWI_TEMPLATE
glob bids-data-dir/dwi-template to find input data
e.g. sub-*/ses-*/dwi/*_dwi.nii.gz (default:
sub-*/dwi/*_dwi.nii.gz)
--t1-template T1_TEMPLATE
glob bids-data-dir/t1-template to find input data e.g.
sub-*/ses-*/anat/*_T1w.nii.gz (default:
sub-*/anat/*_T1w.nii.gz)
--t2-template T2_TEMPLATE
glob bids-data-dir/t2-template to find input data
(default: None)
--task {StructMask,Freesurfer,CnnMask,PnlEddy,FslEddy,TopupEddy,EddyEpi,Ukf,Fs2Dwi,Wmql,Wmqlqc,TractMeasures}
number of Luigi workers
--num-workers NUM_WORKERS
number of Luigi workers (default: 1)
--derivatives-name DERIVATIVES_NAME
relative name of bids derivatives directory,
translates to bids-data-dir/derivatives/derivatives-
name (default: pnlpipe)luigi/scripts/ExecuteTask.py --task Freesurfer --bids-data-dir ~/INTRuST_BIDS -c ~/INTRuST_BIDS/caselist.txt -s ~/INTRuST_BIDS/sessions.txt
firefox http://localhost:8082
Copy the example run_luigi_pnlpipe.lsf script to your directory, edit it, and run as follows:
bsub < /path/to/your/luigi-pnlpipe/workflows/run_luigi_pnlpipe.lsf
NOTE luigid server should be running in the background in a host where all job running hosts can connect to.
bjobs
BIDS specification for naming derivatives is under development and not yet standardized. See here for more details.
- Every derivative file should have a
descfield with relevant value - The file name should have only one suffix that identifies what the file is
such as
_bsefor baseline image,_maskfor mask etc.
derivatives
└── pnlpipe
└── sub-003GNX007
├── anat
│ ├── freesurfer
│ │ ├── label
│ │ ├── mri
│ │ ├── scripts
│ │ ├── stats
│ │ ├── surf
│ │ ├── tmp
│ │ ├── touch
│ │ └── trash
│ ├── sub-003GNX007_desc-T1wXcMabs_mask.nii.gz
│ ├── sub-003GNX007_desc-T2wXcMabs_mask.nii.gz
│ ├── sub-003GNX007_desc-T1wXcMabsQc_mask.nii.gz
│ ├── sub-003GNX007_desc-T2wXcMabsQc_mask.nii.gz
│ ├── sub-003GNX007_desc-Xc_T1w.nii.gz
│ └── sub-003GNX007_desc-Xc_T2w.nii.gz
├── dwi
│ ├── sub-003GNX007_desc-dwiXcEd_bse.nii.gz
│ ├── sub-003GNX007_desc-dwiXcEdMa_bse.nii.gz
│ ├── sub-003GNX007_desc-XcBseBet_mask.nii.gz
│ ├── sub-003GNX007_desc-XcBseBetQc_mask.nii.gz
│ ├── sub-003GNX007_desc-Xc_dwi.bval
│ ├── sub-003GNX007_desc-Xc_dwi.bvec
│ ├── sub-003GNX007_desc-Xc_dwi.nii.gz
│ ├── sub-003GNX007_desc-XcEd_dwi.bval
│ ├── sub-003GNX007_desc-XcEd_dwi.bvec
│ ├── sub-003GNX007_desc-XcEd_dwi.nii.gz
│ ├── sub-003GNX007_desc-XcEdEp_dwi.bval
│ ├── sub-003GNX007_desc-XcEdEp_dwi.bvec
│ └── sub-003GNX007_desc-XcEdEp_dwi.nii.gz
├── fs2dwi
│ ├── eddy_fs2dwi
│ │ ├── b0maskedbrain.nii.gz
│ │ ├── b0masked.nii.gz
│ │ ├── wmparcInBrain.nii.gz
│ │ └── wmparcInDwi.nii.gz
│ └── epi_fs2dwi
│ ├── b0maskedbrain.nii.gz
│ ├── b0masked.nii.gz
│ ├── wmparcInBrain.nii.gz
│ └── wmparcInDwi.nii.gz
└── tracts
├── sub-003GNX007_desc-XcEdEp.vtk
├── sub-003GNX007_desc-XcEd.vtk
├── wmql
│ ├── eddy
│ └── epi
└── wmqlqc
├── eddy
└── epi
To facilitate external collaborators to use luigi-pnlpipe without having to set up a Luigi server at their end, we have set up a public Luigi server. Our server not only runs Luigi tasks but also provides data provenance associated with each output. It is particularly helpful when Luigi tasks are run inside pnlpipe-containers. However, you need a username and password to log in. Please email tbillah@bwh.harvard.edu to obtain that.
- Luigi dashboard https://pnlservers.bwh.harvard.edu/static/visualiser/index.html
- History by task name https://pnlservers.bwh.harvard.edu/history/by_name/Freesurfer
- History by task parameters https://pnlservers.bwh.harvard.edu/history/by_params/Freesurfer?data={"id":"1004"}
- History by task ID https://pnlservers.bwh.harvard.edu/history/by_id/69
- ~ List of all tasks https://pnlservers.bwh.harvard.edu/tasklist
- API graph https://pnlservers.bwh.harvard.edu/api/graph
~ Click on a task to see status, duration, and occasion of all its past runs.
In addition, *.log.html and *.log.json provenance files are generated for each output file in the directory of that file:
sub-1004_ses-01_desc-XcMaN4_T1w.log.html sub-1004_ses-01_desc-Xc_T1w.log.html
sub-1004_ses-01_desc-XcMaN4_T1w.log.json sub-1004_ses-01_desc-Xc_T1w.log.json
sub-1004_ses-01_desc-XcMaN4_T1w.nii.gz sub-1004_ses-01_desc-Xc_T1w.nii.gz
sub-1004_ses-01_desc-XcMaN4_T2w.log.html sub-1004_ses-01_desc-Xc_T2w.log.html
sub-1004_ses-01_desc-XcMaN4_T2w.log.json sub-1004_ses-01_desc-Xc_T2w.log.json
sub-1004_ses-01_desc-XcMaN4_T2w.nii.gz sub-1004_ses-01_desc-Xc_T2w.nii.gzA provenance file captures information about the origin of the output file it is associated with, the parameters and the versions of various software used in the pipeline to generate that file.
Before running any workflow, set the environment variable LUIGI_CONFIG_PATH from where optional parameter values
could be obtained:
export LUIGI_CONFIG_PATH=/path/to/your/pipe_params.cfg
See example parameter files in luigi-pnlpipe/scripts/params/*cfg. You should copy appropriate parameter file (s) among
struct_pipe_params.cfg, dwi_pipe_params.cfg, fs2dwi_pipe_params.cfg and edit that. Finally, set the LUIGI_CONFIG_PATH
variable to the path of your edited parameter file.
# MABS masking of T1w image for case 003GNX007, session BWH01
exec/ExecuteTask --task StructMask \
--bids-data-dir ~/Downloads/INTRuST_BIDS -c 003GNX007 -s BWH01 --t1-template sub-*/anat/*_T1w.nii.gz
# MABS masking of T2w image for all cases and all sessions
exec/ExecuteTask --task StructMask \
--bids-data-dir ~/Downloads/INTRuST_BIDS \
-c ~/Downloads/INTRuST_BIDS/caselist.txt \
-s ~/Downloads/INTRuST_BIDS/sessions.txt \
--t2-template sub-*/anat/*_T2w.nii.gz \
--num-workers 3
# Freesurfer segmentation using only T1w image for case 003GNX007
exec/ExecuteTask --task Freesurfer \
--bids-data-dir ~/Downloads/INTRuST_BIDS -c 003GNX007 --t1-template sub-*/anat/*_T1w.nii.gz
# Freesurfer segmentation using both T1w and T2w image for all cases
exec/ExecuteTask --task Freesurfer \
--bids-data-dir ~/Downloads/INTRuST_BIDS \
-c ~/Downloads/INTRuST_BIDS/caselist.txt \
-s BWH01 \
--t1-template sub-*/anat/*_T1w.nii.gz --t2-template sub-*/anat/*_T2w.nii.gz \
--num-workers 3
# pnl_eddy.py requires only DWI image
exec/ExecuteTask --task PnlEddy \
--bids-data-dir ~/Downloads/INTRuST_BIDS -c 003GNX007 --dwi-template sub-*/dwi/*_dwi.nii.gz
# pnl_epi.py requires a DWI and a T2w image
exec/ExecuteTask --task PnlEddyEpi
--bids-data-dir ~/Downloads/INTRuST_BIDS -c ~/Downloads/INTRuST_BIDS/caselist.txt \
--t2-template sub-*/anat/*_T2w.nii.gz --dwi-template sub-*/dwi/*_dwi.nii.gz \
--num-workers 3
# UKFTractography based on pnl_eddy.py corrected data, requires only DWI image
exec/ExecuteTask --task Ukf
--bids-data-dir ~/Downloads/INTRuST_BIDS -c ~/Downloads/INTRuST_BIDS/caselist.txt \
--dwi-template sub-*/dwi/*_dwi.nii.gz \
--num-workers 3
# UKFTractography based on pnl_epi.py corrected data, requires a DWI and a T2w image
exec/ExecuteTask --task Ukf
--bids-data-dir ~/Downloads/INTRuST_BIDS -c ~/Downloads/INTRuST_BIDS/caselist.txt \
--t2-template sub-*/anat/*_T2w.nii.gz --dwi-template sub-*/dwi/*_dwi.nii.gz \
--num-workers 3
# Wmqlqc based on pnl_eddy.py corrected data, requires only DWI image
exec/ExecuteTask --task Wmqlqc
--bids-data-dir ~/Downloads/INTRuST_BIDS -c 003GNX007 --dwi-template sub-*/dwi/*_dwi.nii.gz
# Wmqlqc based on pnl_epi.py corrected data, requires a DWI and a T2w image
exec/ExecuteTask --task Wmqlqc
--bids-data-dir ~/Downloads/INTRuST_BIDS -c ~/Downloads/INTRuST_BIDS/caselist.txt \
--t2-template sub-*/anat/*_T2w.nii.gz --dwi-template sub-*/dwi/*_dwi.nii.gz \
--num-workers 3
This section is for an experienced Python programmer who can debug issues and potentially contribute to
luigi-pnlpipe. For convenience of the user, all tasks are run from one script: exec/ExecuteTask.
However, these tasks are indeed defined in separated modules:
~/luigi-pnlpipe/workflows/struct_pipe.py
~/luigi-pnlpipe/workflows/dwi_pipe.py
~/luigi-pnlpipe/workflows/fs2dwi_pipe.py
To know which tasks are defined in each of the modules, first set the PYTHONPATH and then do:
cd ~/luigi-pnlpipe
export PYTHONPATH=`pwd`/scripts:`pwd`/workflows:$PYTHONPATH
# to view all tasks and their respective parameters, same for dwi_pipe and fs2dwi_pipe
luigi --module struct_pipe --help-all
# to view parameters of one task, same for dwi_pipe and fs2dwi_pipe
luigi --module struct_pipe StructMask --help
You should provide value for each of the arguments or at least the mandatory ones:
luigi --module struct_pipe StructMask
--id ID
--ses SES
--bids-data-dir BIDS_DATA_DIR
--struct-template STRUCT_TEMPLATE
--derivatives-dir DERIVATIVES_DIR
--csvFile CSVFILE
--debug
--fusion FUSION
--mabs-mask-nproc MABS_MASK_NPROC
--ref-img REF_IMG
--ref-mask REF_MASK
--reg-method REG_METHOD
--slicer-exec SLICER_EXEC
--mask-qc
ipython
from struct_pipe import StructMask
from luigi import build
build([StructMask(id= '001',
ses= 'BWH01',
bids_data_dir= '/home/tb571/Downloads/INTRUST_BIDS',
struct_template= 'sub-*/anat/*_T2w.nii.gz',
csvFile= '-t2',
debug= False,
fusion= 'avg',
mabs_mask_nproc= 8,
ref_img= '',
ref_mask= '',
reg_method= '',
slicer_exec= '',
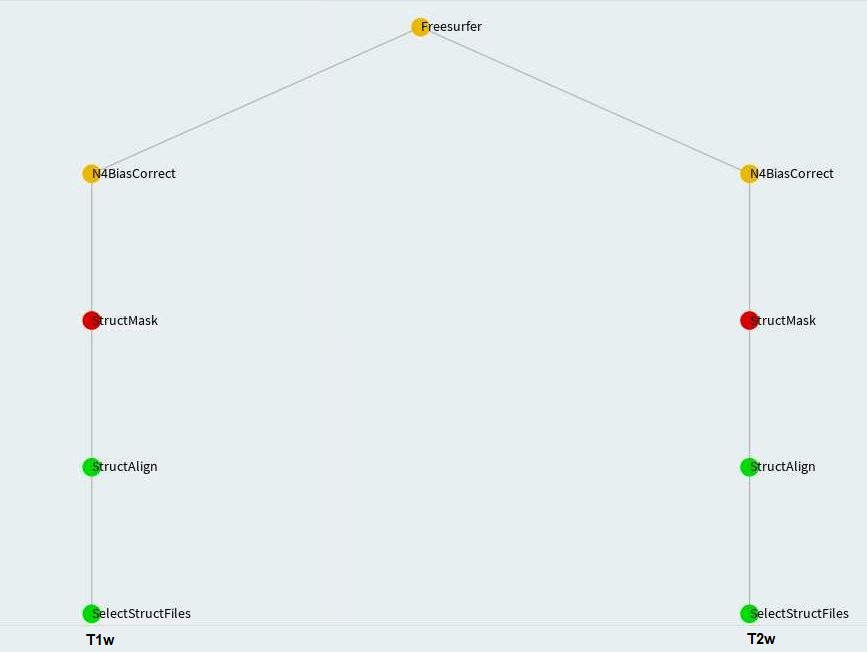
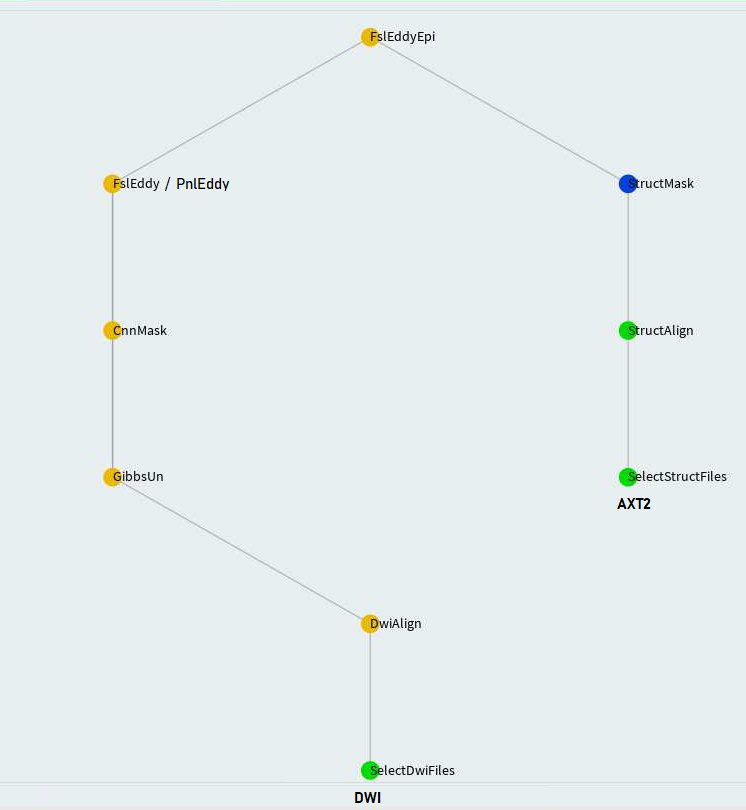
mask_qc= True)])FslEddy and PnlEddy tasks are represented by the left branch in the above. Again, when PnlEddy is used, the task name becomes PnlEddyEpi.
When two opposing acquisitions--AP and PA are available, eddy+epi correction can be done in a more sophisticated way through TopupEddy.
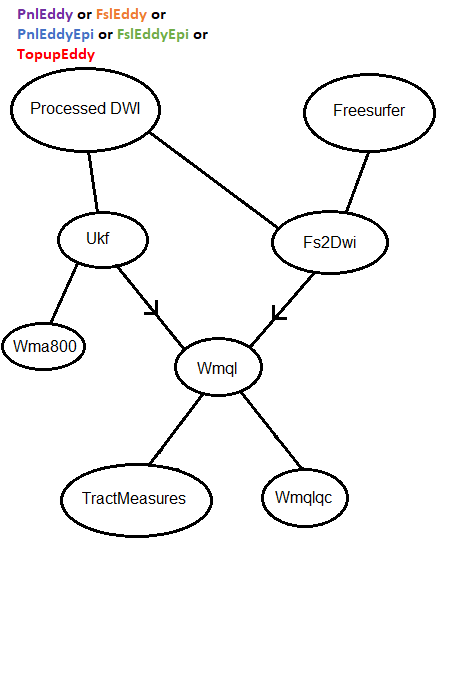
Additionally, some more complicated flowcharts can be found here.
Mandatory parameters are provided from command line with exec/ExecuteTask script.
--bids-data-dir BIDS_DATA_DIR
/path/to/bids/data/directory
-c C a single case ID or a .txt file where each line is a
case ID
-s S a single session ID or a .txt file where each line is
a session ID (default: 1)
--dwi-template DWI_TEMPLATE
glob bids-data-dir/dwi-template to find input data
e.g. sub-*/ses-*/dwi/*_dwi.nii.gz (default:
sub-*/dwi/*_dwi.nii.gz)
--t1-template T1_TEMPLATE
glob bids-data-dir/t1-template to find input data e.g.
sub-*/ses-*/anat/*_T1w.nii.gz (default:
sub-*/anat/*_T1w.nii.gz)
--t2-template T2_TEMPLATE
glob bids-data-dir/t2-template to find input data
(default: None)
--task {StructMask,Freesurfer,CnnMask,PnlEddy,PnlEddyEpi,FslEddy,FslEddyEpi,TopupEddy,Ukf,PnlEddyUkf,Fs2Dwi,Wmql,Wmqlqc}
number of Luigi workers (default: None)
Each of the luigi-pnlpipe/scripts/*py modules have optional parameters. The optional parameters are equipped with
default values such that you would be able to run luigi-pnlpipe without having to modify any of the three files:
struct_pipe_params.cfg, dwi_pipe_params.cfg, and fs2dwi_pipe_params.cfg. Whether you need to edit the parameter
files or not, see the beginning of Example Commands section to know how a parameter file is
interfaced with luigi-pnlpipe.
The three parameter files are shown here. The parameters defined in them, bear consistency with that of individual modules.
As usual, you can check individual module parameters as luigi-pnlpipe/scripts/atlas.py --help-all and so on.
Used by StructMask, N4BiasCorrect and Freesurfer tasks.
Let's say you want to create a mask for T2w image of a subject where you have already obtained T1w image from atlas.py.
Given T1w and T2w images were obtained through same protocol, you no longer need to do resource consuming atlas.py for
T2w again. Rather, you can perform a rigid ANTs registration with T1w image and mask as reference. For this shortcut,
you would edit the following parameters in struct_pipe_params.cfg:
[StructMask]
ref_img: /path/to/T1w/image
ref_mask: /path/to/T1w/atlas/maskThe StructMask and BseBetMask tasks come with quality checking feature enabled. They do not do the quality checking
automatically, but let you do it and advance from there. If you set:
slicer_exec: /path/to/a/valid/slicer
Then, the pipeline will load the created mask overlaid on the structural/baseline image for you to check quality.
After you check the mask or potentially edit it, save it with Qc at the end of the value in _desc field. A direction
will already be available on your console:
** Check quality of created mask /tmp/sub-003GNX007_desc-T1wXc_mask.nii.gz .
Once you are done, save the (edited) mask as /tmp/sub-003GNX007_desc-T1wXcQc_mask.nii.gz **
There is one limitation with the above integration. You need to be in front of the machine where you are creating the mask.
So, it is feasible on a workstation or an individual HPC node through NoMachine. However, if you use bsub to run a job,
you will not have GUI support. In that case, you can use:
mask_qc: True
Like the above, after mask is created, the program will poll the file system for /tmp/sub-003GNX007_desc-T1wXcQc_mask.nii.gz.
Once file is obtained, the pipeline will progress from there. But unlike the function triggered by slicer_exec, it will
not attempt to load the mask on a visualizer. After you see the above direction on your console, you can quality check
the mask using your favorite visualizer and save it like shown above.
Used by StructMask, BseBetmask, BseExtract, PnlEddy, FslEddy, EddyEpi, TopupEddy, Ukf, and Wma800 tasks.
Each parameter is preceded by the task names that use the parameter. You may see Workflows to know which requisite tasks are run as part of the task you want to run. Then, you should edit only the parameters relevant to your task(s).
Used by Fs2Dwi, Wmql, Wmqlqc, and TractMeasures tasks.
-
To force re-running of a job, please delete all intermediate outputs. See how Luigi works for details.
-
Use appropriate parameter file to define
LUIGI_CONFIG_PATH. Please edit relevant parameters only. -
If you attempt to re-run a job, but:
- it does not get run permission
- or shows as being executed by another scheduler
You should:
- mark that job as done on the visualizer
- or kill and restart
luigidserver to re-run successfully
To kill luigid server, use killall -9 luigid.
- There are two types of Dependency Graph:
SVGandD3. You can switch between them by clicking on the tab on top right corner. When the parent task has a bunch of dependencies,SVGgraph depicts the order in which jobs are picked up for running. On the other hand,D3graph shows the actual dependency. See luigi/issues/2889 for better explanation. So, it is advised to always look atD3graph.
Feel free to report any other issues at https://github.com/pnlbwh/luigi-pnlpipe/issues We shall get back to you at our earliest convenience.