mkCOInr - COInr
mkCOInr is a series of Perl scripts that aims to create and customize COInr, a large, comprehensive, COI database form NCBI-nucleotide and BOLD.
The COInr database is composed of two files
- COInr.tsv contains sequenceIDs, taxIDs, and sequences
- taxonomy.tsv contains all taxIDs and associated information
COInr is freely available and can be easily downloaded at
Documentation
The mkCOInr documentation is hosted at ReadTheDocs.
COInr
- COInr is not specific to a particular region of the COI gene (sequences can be partial).
- All cellular organisms are included, even Bacteria.
- Sequences with incomplete lineages (e.g. assigned to a family without further precision) are present in the database
- Taxa are taken into account only with correct latin name formats (e.g. instead of 'Proterorhinus sp. BOLD:EUFWF4948-19', the sequence is assigned to Proterorhinus genus without a species name)
The database can be used directly for similarity-based taxonomic assignations of metabarcoding data with any COI marker (primer pairs) of any geographical regions or target group.
Alternatively, the database can be used as a starting point to create smaller, more specific custom databases.
mkCOInr
Sequences from COInr can be selected for :
- A particular gene region (amplicon of a given primer pair)
- List of taxa (sequences of a taxon list can be eiter selected or eliminated)
- User-defined minimal taxonomic resolution
This can save a considerable amount of time and effort, since one of the most important challenges of creating a custom database is the mass downloading of the sequences and their pooling into a coherent taxonomic system.
Custom sequences can also be included and their taxonomic lineages is correctly handled.
COInr or the custom databases derived from it can be formated to different database formats (qiime, rdp, blast, vtam, full).
Fig.1 The full pipeline to create COInr and options to make a custom database
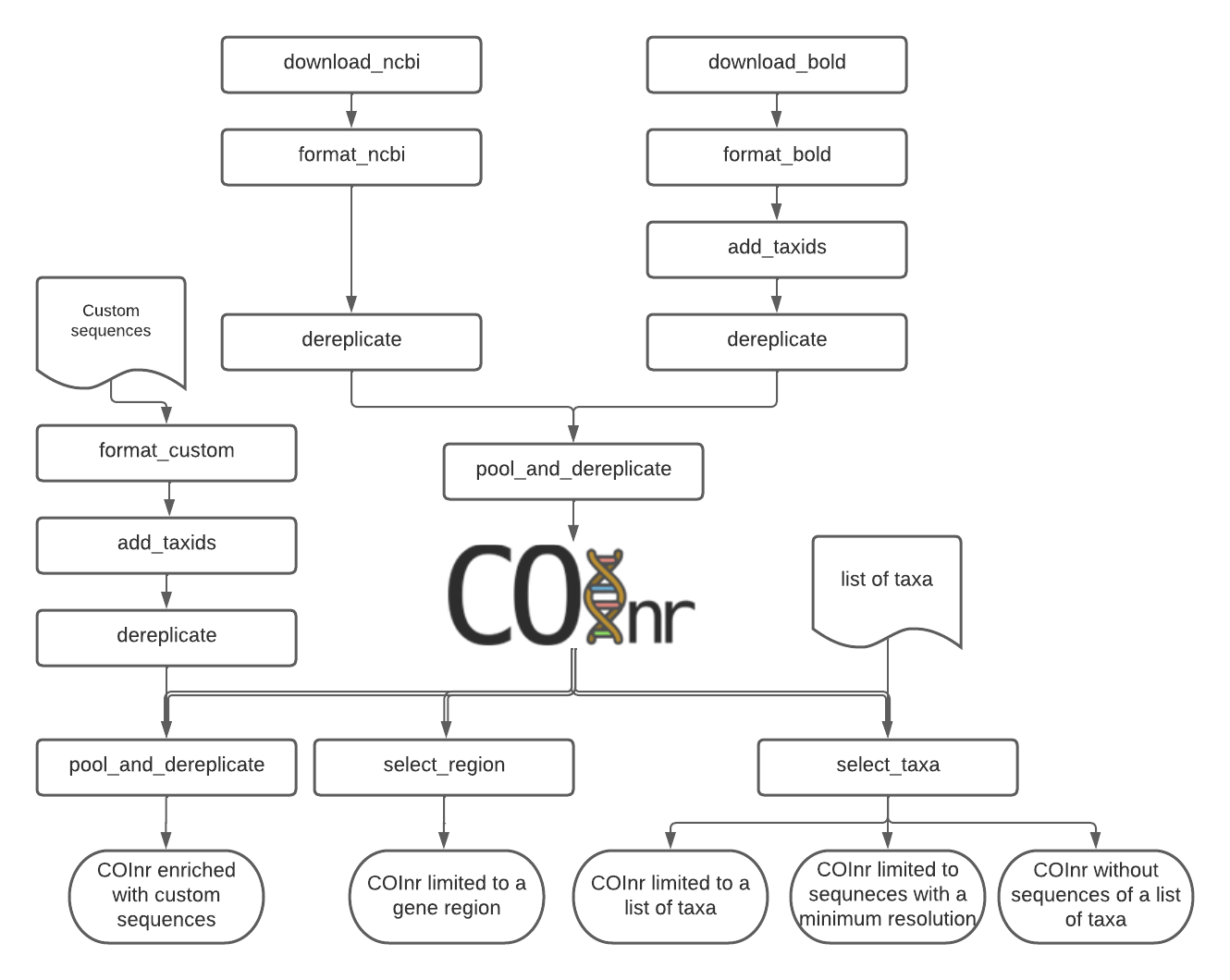
Major features of the creation of COInr
- Mass download of sequences and their taxonomic lineages from NCBI-nucleotide and BOLD databases
- TaxIDs are used to avoid problems with homonyms and synonyms
- Creation of a coherent taxID system. The hierarchical structure of the NCBI taxIDs is completed if necessary with new, negative taxIDs.
- When adding sequences with unknown taxIDs, taxon names are matched to already existing taxonomic lineages in the database to identify a correct existing taxID, or to assign a new one.
- Taxonomically aware demultiplexing
- Creation of a ready-to-use database in BLAST, RDP_classifier QIIME or a FULL tsv format
Warning
- The taxonomic origin of the sequences is not checked, but taken as a face value from the source database.
- At the scale of the complete database I did not find a satisfactory method to blacklist sequences that are probably incorrectly assigned. However, if a small custom database is produced, the use of a phylogenetic method like SATIVA (https://github.com/amkozlov/sativa) becomes feasible and recommended to eliminate sequences of dubious origin.