Conway's Game of Life implementations
Just playing around, this is in no way complete and likely never will be. :)
Plans:
- Java implementation: This was the first implementation. The game logic is complete, but no work on user interface.
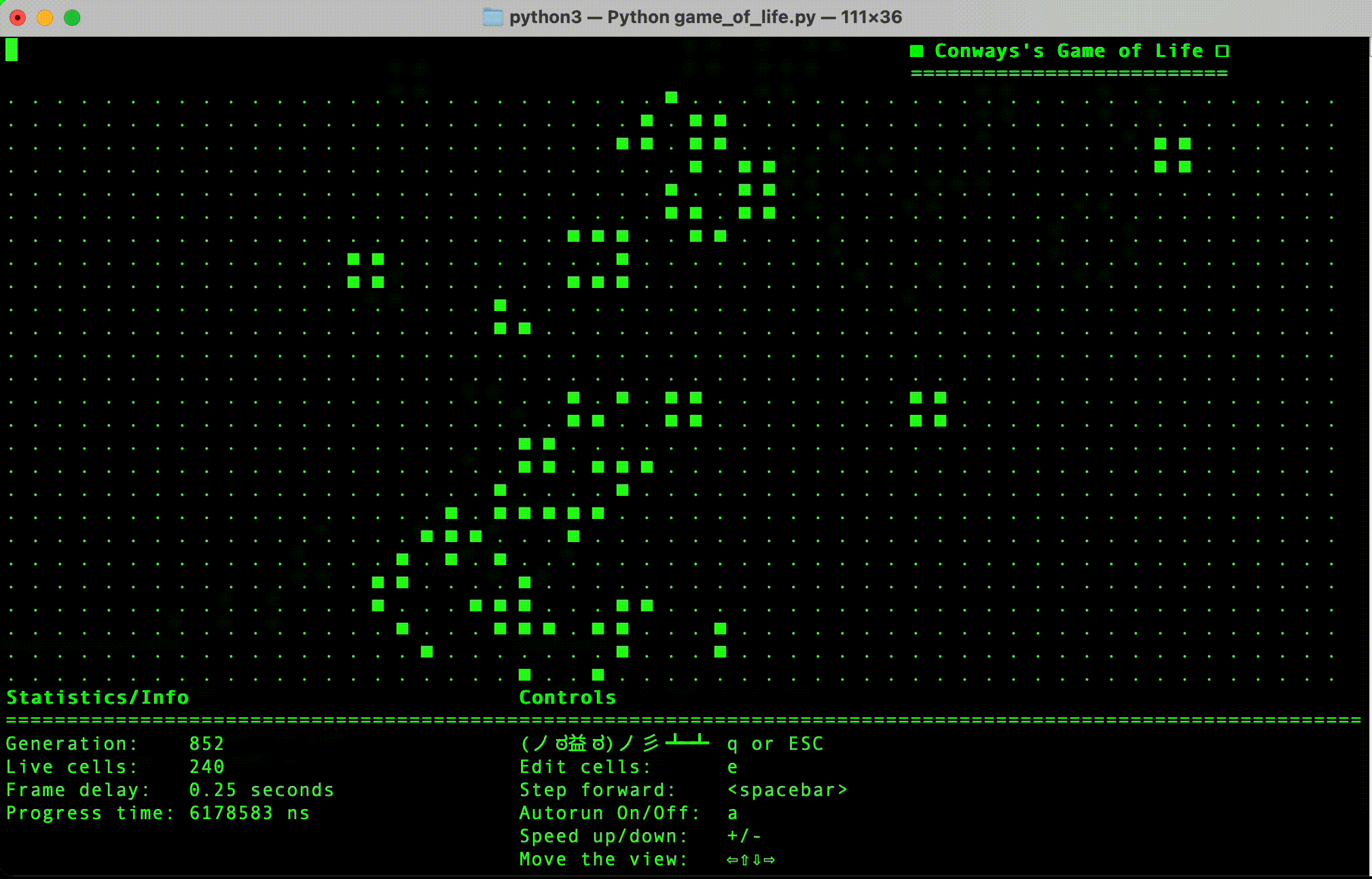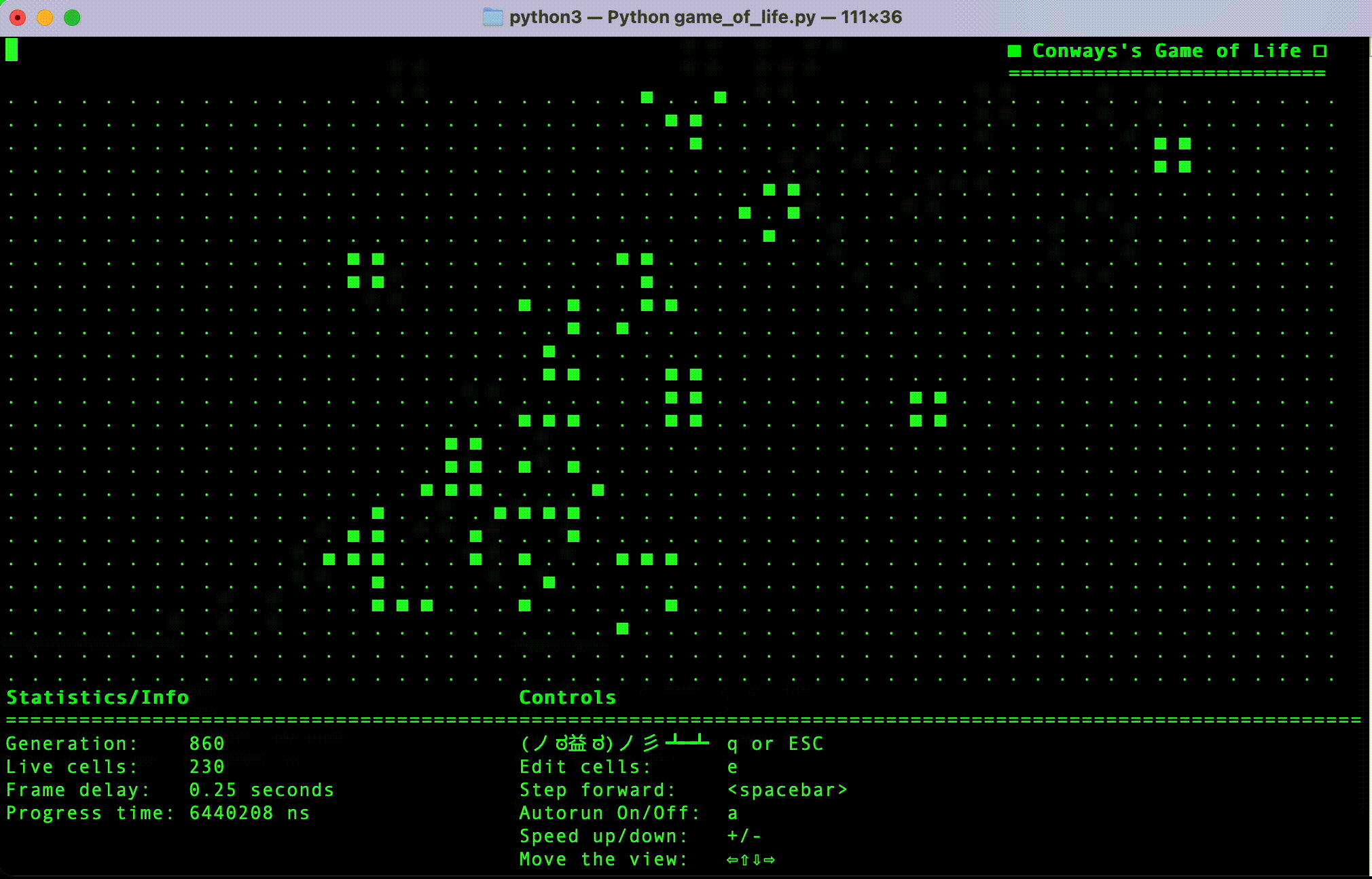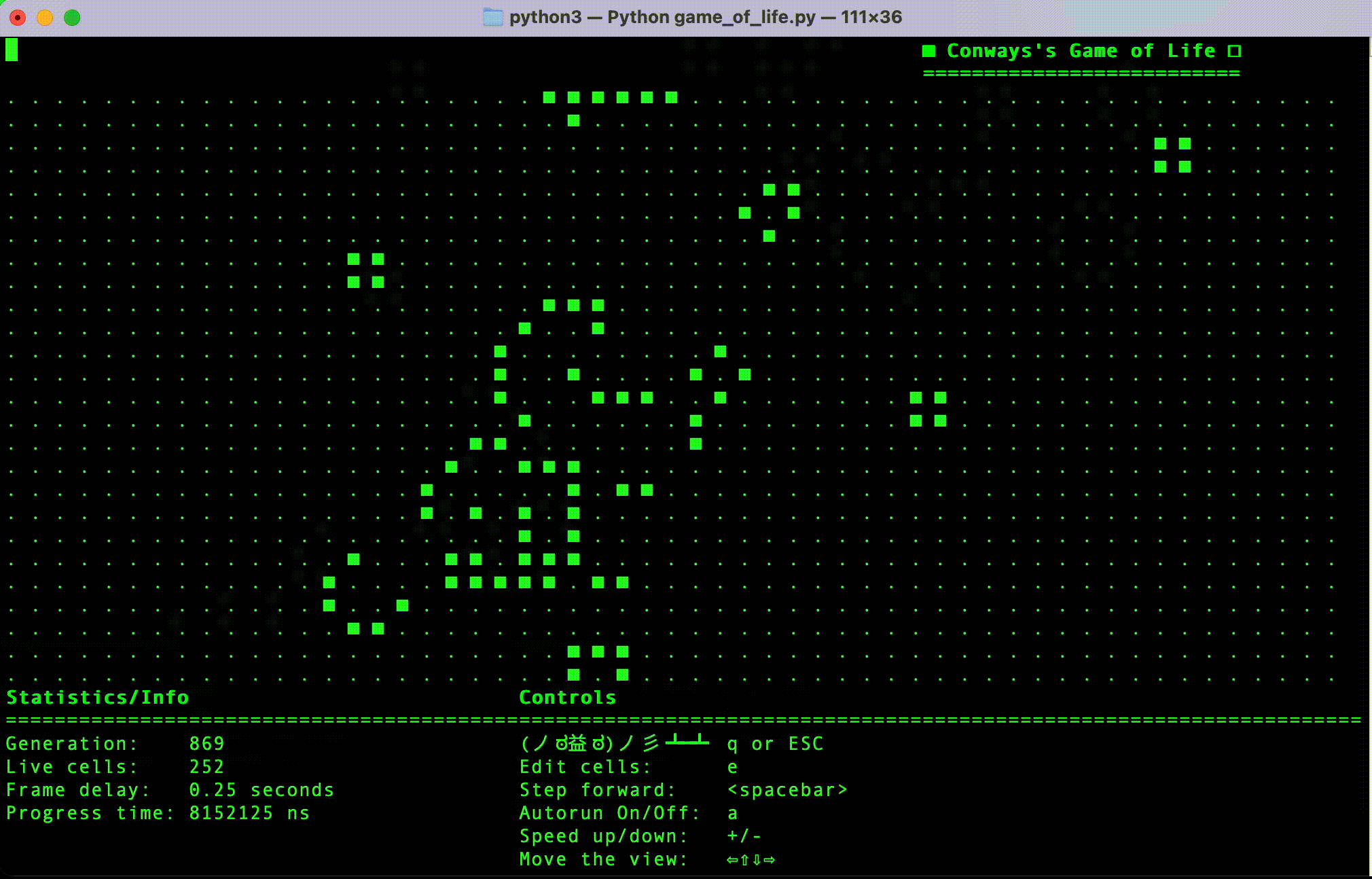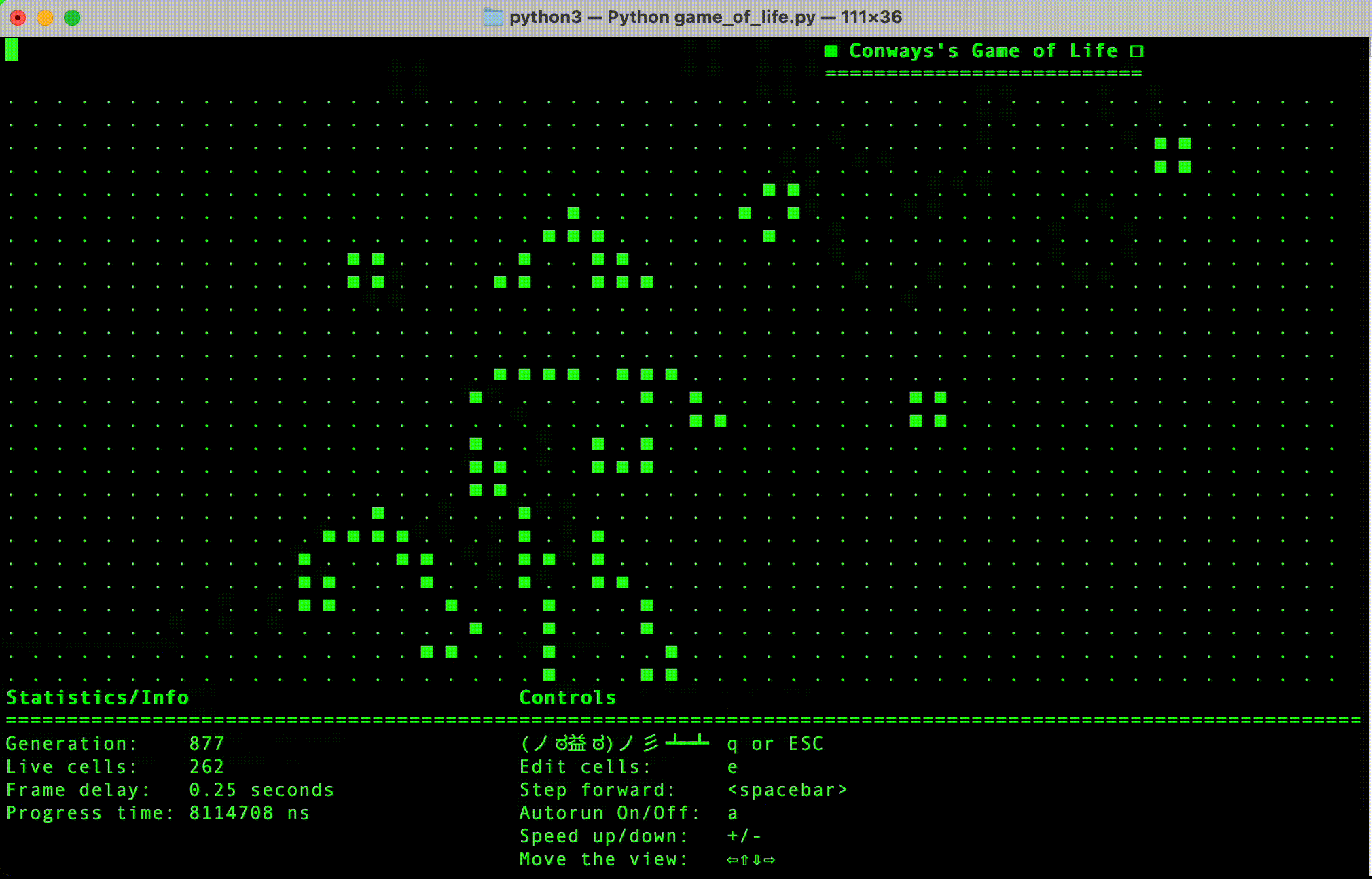
- Python implementation: This is currently the most progressed, and has a simple terminal user interface.
- TypeScript in NodeJS implementation: Planned, currently unstarted.
- Haskell implementation: Yeah maybe, would be interesting.
- C# implementation: Maybe.
TODO:
- Some form of different UI allowing different zoom levels? (Preferably not a web interface...)
- Add ability to use different rules.
- Ability to get RLE and other formats directly from (popular?) GoL sites on the internet.
- Ability to reset back to last edit. Probably implemented that we save the state when we exit Edit mode.
- (Done for Python) Ability to write out the current board to a file, "Save Game" function.
- (Done for Python) Add other formats such as Plaintext.
- (Done for Python) Add RLE (Run Length Encoded) file format reading.
- (Done for Python) GitHub Actions CI?
- (Done for Python) Codecov?
- (Done in Python) Implement simple terminal based interface.
- (Done in Java, Python) Add GitHub Dependabot.
- (Done in Java, Python) Add tests.
- (Done in Java, Python) Implement game logic.
- (Done in Java, Python) Use languange appropriate build management tools.
Conway's Game of Life rules
Original Rules:
- Any live cell with fewer than two live neighbours dies, as if by underpopulation.
- Any live cell with two or three live neighbours lives on to the next generation.
- Any live cell with more than three live neighbours dies, as if by overpopulation.
- Any dead cell with exactly three live neighbours becomes a live cell, as if by reproduction.
Condensed Rules:
- Any live cell with two or three live neighbours survives.
- Any dead cell with three live neighbours becomes a live cell.
- All other live cells die in the next generation. Similarly, all other dead cells stay dead.
Further simplified Rules that are easier to program:
- Any cell, dead or alive, with exactly 3 neighbours is alive in the next generation.
- A live cell with exactly 2 neighbours is alive in the next generation.
- All other cells are dead in the next generation.
Approaches
Set based simple implementation
- Create new Set<int, int>,
A. This set simply records the coordinates as (row,col) of live cells. By being in the set it's a live cell. We don't record dead cells, so it is a very sparse data structure. - Create new set
Bfor next generation. - Create a new ephemeral set for recording dead cells that have been checked this generation,
tmp. - Iterate through all live cells in
A- For each cell, check how many of the 8 neighbours are alive in
A. If 2 or 3, this cell stays alive so add the coordinates to the next-genBset. - Get the collection of the 8 neighbours that are dead.
- Check each of the dead cells if it has already been checked by looking if it is in
tmp. - Check if the dead neighbour has exactly 3 live neighbours in
A, in which case it will come alive in the next generation, so add it toBmeaning it will be live. - Record the dead cell in
tmpas having been checked. - We have now checked all live cells, and all their neighbours - this means we have checked all cells in the universe that could possibly become live.
- For each cell, check how many of the 8 neighbours are alive in
- At this point the next generation of live cells is ready in
B. - Assign
BtoA, cleartmp.
Negatives:
- only 2 states per cell - alive (coordinates present) or dead (coordinates not present)
- does a lot of set lookups:
- 8 lookups to find the neighbours of each live cell
- 1 lookup for each dead neighbour to see if it has already been checked
- for each of the dead neighbours that has not already been checked, another 8 lookups to see if it has enough live neighbours to come alive
Positives:
- memory efficient as we only store coordinates, no other state
- able to create much larger universes compared to e.g. arrays or the map implementation - as long as the universe is sparsely populated
- no edge to the universe except for the languages maximum key value (e.g. in Java Integer.MAX) which gliders etc will eventually hit - could try to deal with this by using some implementation of e.g. BigInteger?
- not too hard to understand
2D arrays
A[rows][columns]array created at init- Set each individual live cell in
A - Create parallel
Barray set to same size but all cells are dead - Algorithm loops through each element
A[row][col]- Test the 8 adjacent elements of
A[row][col], save the result inB[row][col] - When testing, overflow on edges so if testing element at edges
cols.maxorrows.max, overflow and test from opposite edge. Same forcolumns.minandrows.min
- Test the 8 adjacent elements of
- Assign
BtoA - Print
A
Negatives:
- need
rows*columns*2of memory, which limits max size - we test each element so
O(rows*columns), also limits max size
Positives:
- it's fast for small sized boards
- no hashtable lookups
- simple to understand
TreeMap simple implementation - actually scratch that, the Python version now uses a normal map/dict for better performance.
- Create new TreeMap<int[], Cell>, A.
- The key is the int[row,col] of the Cell.
- We rely on the ordering of the keys when later iterating to create an "array view" for display.
- Funnily enough the way the python version was implemented this is not actually true and we do not rely on the ordering. Changing to a normal dict resulted in an immediate 30% performance increase.
- Revisit this in the Java version some time.
- Need to check what natural ordering is of int[row,col] - might have to provide our own Comparator. When we iterate over the keys they need to come out in row order left to right.
- Create new live Cells and put/replace in map at their row,col coordinate. No need to check if it exists already.
- Check in map if each neighbour exists. For each that does not exist, add a new dead Cell.
- Now we have all live and dead Cells that need to be considered when progressing a generation.
- Create new map B for next generation.
- Iterate through all Cells in A
- Check each cell - both live and dead - if it will be alive in next gen by doing lookups for all 8 of its neighbours in A. Use predefined
int[row,col]for N, NE, E, SE, S, SW, W, NW neighbours to avoid garbage collection? Maybe test this for speed. - Because we have also checked all dead neighbours of all live cells, we have checked all cells in the universe that could possibly become live.
- If it will be alive, create a new Cell in B, with all it's dead neighbours as above.
- If it will be dead, do not add to B.
- Check each cell - both live and dead - if it will be alive in next gen by doing lookups for all 8 of its neighbours in A. Use predefined
- At this point, next generation is ready in B, and has only the live Cells and their immediate dead neighbours.
- Assign B to A.
Negatives:
- does a lot of map lookups
Positives:
- able to create much larger universes compared to arrays - as long as the universe is sparsely populated
- no edge to the universe except for Integer.MAX for keys which gliders etc will eventually hit - could try to deal with this by using BigInteger?
- not too hard to understand