Unconstrained lattice antibody-antigen bindings generator - One tool to simulate them all!
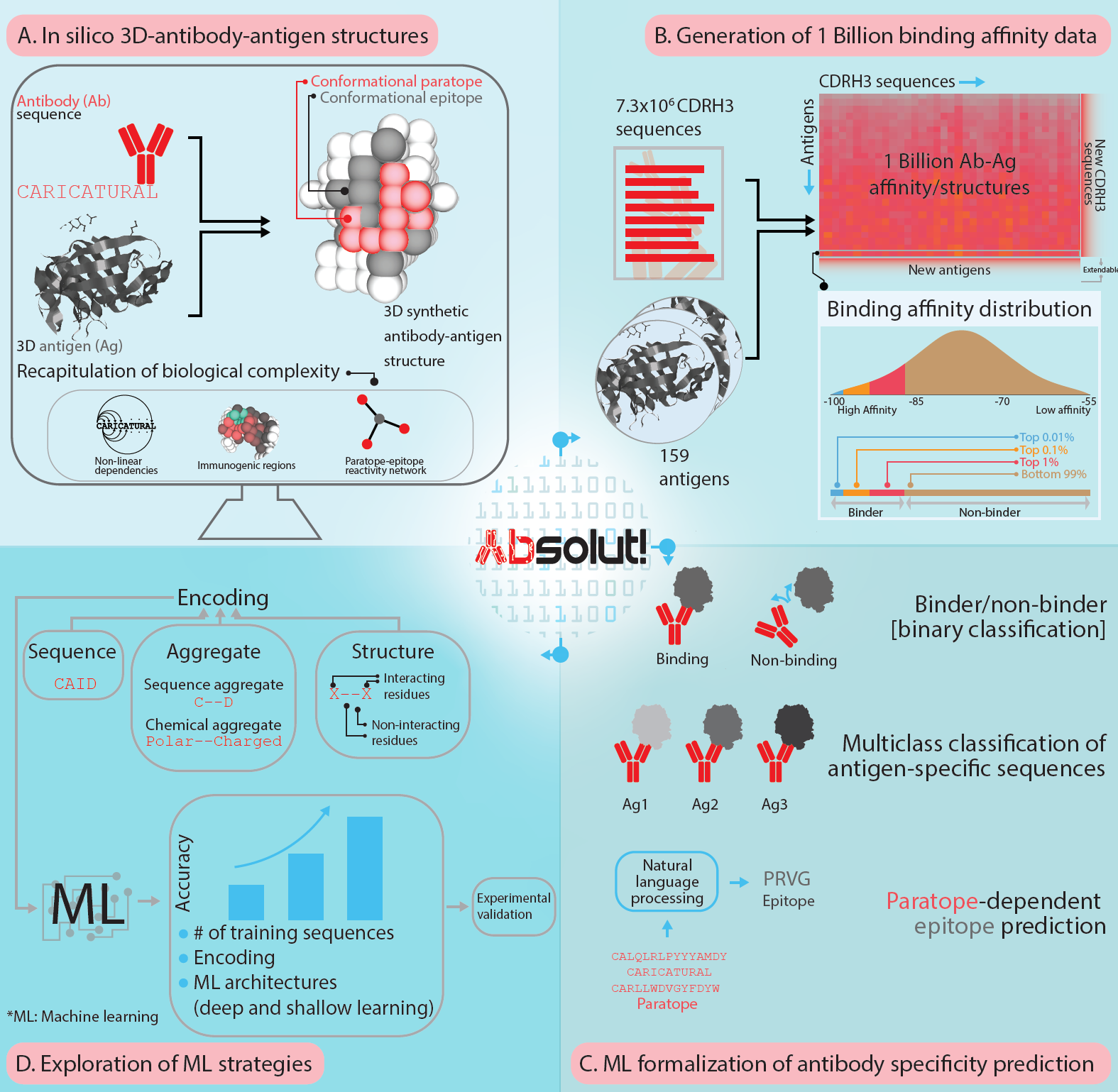
Absolut! allows the user to generate custom datasets by discretizing new antigens or annotating custom CDRH3 sequences enabling the on-demand generation of large synthetic datasets for 3D-antibody-antigen binding generated under the same, deterministic settings, with accessible computing resources.
A Greifflab product!
Absolut! software
- takes PDB antigens, converts them into lattice representation with integer positions.
- takes CDR3 Amino Acid sequences, and computes their best binding around a lattice antigen.
- generates features of antibody-antigen bindings directly usable for Machine Learning
Absolut! database
The database generated with Absolut! is available as repository in NIRD research data storage. The files can directly be accessed by http Database Files.
Absolut! is distributed under the GNU affero general public license (see License.md)
You can find a more detailed documentation in the subfolder doc/HowToAbsolut.pdf
The method and analyses of antibody-antigen bindings are explained in:
- Robert and Akbar et al. 2021, 'A billion synthetic 3D-antibody-antigen complexes enable unconstrained machine-learning formalized investigation of antibody specificity prediction', BioRXiV link
- Robert et al 2020, 'Ymir: A 3D structural affinity model for multi-epitope vaccine simulations', BioRXiV link
It has been used to benchmark antibody generative ML models in: Akbar and Robert et al 2021, 'In silico proof of principle of machine learning-based antibody design at unconstrained scale' link
git clone git://github.com/csi-greifflab/AbsolutAbsolut! can be compiled into three versions: one that doesn't require any library to be installed (AbsolutNoLib), the full version (Absolut), and a version specific for using MPI (AbominationMPI). See Installation for details.
Calling Absolut alone shows the list of available options.
./Absolut
#or
./AbsolutNoLibAn all-in-one example for most tasks is:
plan 3GB of disk space, 2 GB memory and 500MB download
#With ./Absolut or ./AbsolutNoLib:
# Get the list of available lattice antigens
./AbsolutNoLib listAntigens
# Get the list of precomputed CDRH3 possible structures for an antigen
./AbsolutNoLib info_fileNames 1FBI_X
wget http://philippe-robert.com/Absolut/Structures/SULSU040643e2c0a6d6343bbe8a27b079ef91-10-11-efc862c2cdef086ba79606103a3dfc62Structures.txt
# Get the antibody-antigen complex of a CDRH3 sequence to an antigen (requires the downloaded file above to be in the same folder)
./AbsolutNoLib singleBinding 1FBI_X CARAAHKLARIPK
# High throughput calculation of antibody-antigen complexes for a list (repertoire) of CDRH3 sequences. Here, requests 10 threads in parallel.
./AbsolutNoLib repertoire 1FBI_X SmallSetCDR3.txt 10 #SmallSetCDR3.txt is a provided example list of CDR3s inside src/
# Calculation of structure and sequence encodings from the produced list of antibody-antigen complexes
./AbsolutNoLib getFeatures 1FBI_X 1FBI_XFinalBindings_Process_1_Of_1.txt outputFeaturesFile.txt 1 true
# Generating new antigens and visualizing complexes is only possible with the full ./Absolut
# Takes a PDB and the chains of interests, lattice resolution and discretization method and returns the C++ code for the antigen [New antigens would need to be manually added to antigenLib.cpp].
./Absolut discretize 1CZ8 VW 5.25 FuC
# Visualizing of available antigens in 3D
./Absolut visualize 1FBI_X
# Visualizing many binding complexes
./Absolut visualize 1FBI_X 1FBI_XFinalBindings_Process_1_Of_1.txt -95.5
# Visualizing binding hotspots
./Absolut hotspots 1FBI_X 1FBI_XFinalBindings_Process_1_Of_1.txt -95.5 4 Absolut! was written in C++.
Three versions are provided with more or less library requirements: Absolut, AbsolutNoLib and AbominationMPI. All versions require a C++ compiler. We recommend starting with AbsolutNoLib, which is easier to install, as most tasks are already available with this version.
1/ AbsolutNoLib requires no additional library, and can perform all tasks except user interface for discretization and 3D visualization. We recommend first compiling/running AbsolutNoLib, which should work smoothly.
cd src
make # This creates 'AbsolutNoLib' executable. 2/ AbominationMPI is the MPI parallelized version for high throughput repertoire bindings generation.
- requires MPI compiler and headers
cd src
#Depending on your MPI compiler,
make MPIcxx
#or
make MPIc++
#or
make MPIgxx
# This creates 'AbominationMPI' executable.3/ Absolut is the full version,
- requires the Qt framework
- requires freeglut library (or another other C++ glut library) fro visualizing 3D lattice structures
- requires the gsl library for discretizing new antigens
- requires wget, for downloading files (like PDBs when discretizing)
- optionally, a sorftware to visualize PDBs (like Rasmol)
Installing and then linking the libraries can be tricky depending on the OS. Full help for installing these libraries (especially in Windows) is provided in the documentation doc/HowToAbsolut It is also possible to use Absolut! with a user-defined custom subset of libraries (See in the full documentation).
Installing on linux (simplest):
#to find the available packages in your distribution
apt-cache search qtbase
apt-cache search libqt5
#These packages should do the job
sudo apt-get install qtbase5-dev
sudo apt-get install qtcreator
sudo apt-get install libqt5svg5
sudo apt-get install libqt5printsupport5
sudo apt-get install libgsl-devInstalling on Windows:
Compiling
#Please DO NOT compile the full version in src/ folder, it would destroy the Absolut/ subfolder and the original Makefile ...
#We recommend to compile into src/bin because Absolut! expects the pdb-tools scripts to be directly in ../pdb-tools
cd src/bin/
qmake ../Absolut/Absolut.pro #qmake creates a new Makefile embedding Qt libraries locations
make #This creates AbsolutAlternately, if you have installed the Qt framework with qtcreator,
qtcreator Absolut/Absolut.pro
#or
qtcreator Absolut/AbsolutNoLib.pro
#or
qtcreator Absolut/AbsolutNoLibMPI.proInstalling on MAC:
#if no recent g++ compiler, use this command (will also install g++)
brew install gcc
brew install wget
brew install qt
brew install gsl
brew install freeglutThen, from the src/bin/ folder,
qmake ../Absolut/Absolut.pro
makeSome OS specific points: Inside Absolut/Absolut.pro, your compiler might recognize only -o1 or -O1. Linking the libraries might require that you provide their folder. Can either add it in the Absolut.pro linker options, or inside the Makefile manually (but then do not run qmake anymore). -framework openGL should work for openGL and already be added. Depending on your g++ compiler, there might be conflicts between the C++ language of the libraries and the C++ standard libraries provided with the compiler. This might be solved by adding "QMAKE_CXXFLAGS += -std=c++14 -std=c++17" inside Absolut/Absolut.pro. We didn't add these lines by default in case your C++ compiler doesn't support C++17.
Note, the libraries Latfit, pdb-tools and soil are provided inside Absolut with their compatible version (and do not need to be installed by the user) - Latfit has been slighty modified to consider the center of a full residue (not only the side-chain) during discretization
AbsolutNoLib has been run on a HPC cluster with CentOS Linux version 7, and gcc version 4.8.5 (no other libraries required). The full Absolut! has been run on windows 10, using gcc 7.3.0 (MinGW-W64), gsl 2.6, GNU Wget 1.11.4, Qmake 3.1, Qt version 5.12.5 and freeglut 3.2.1
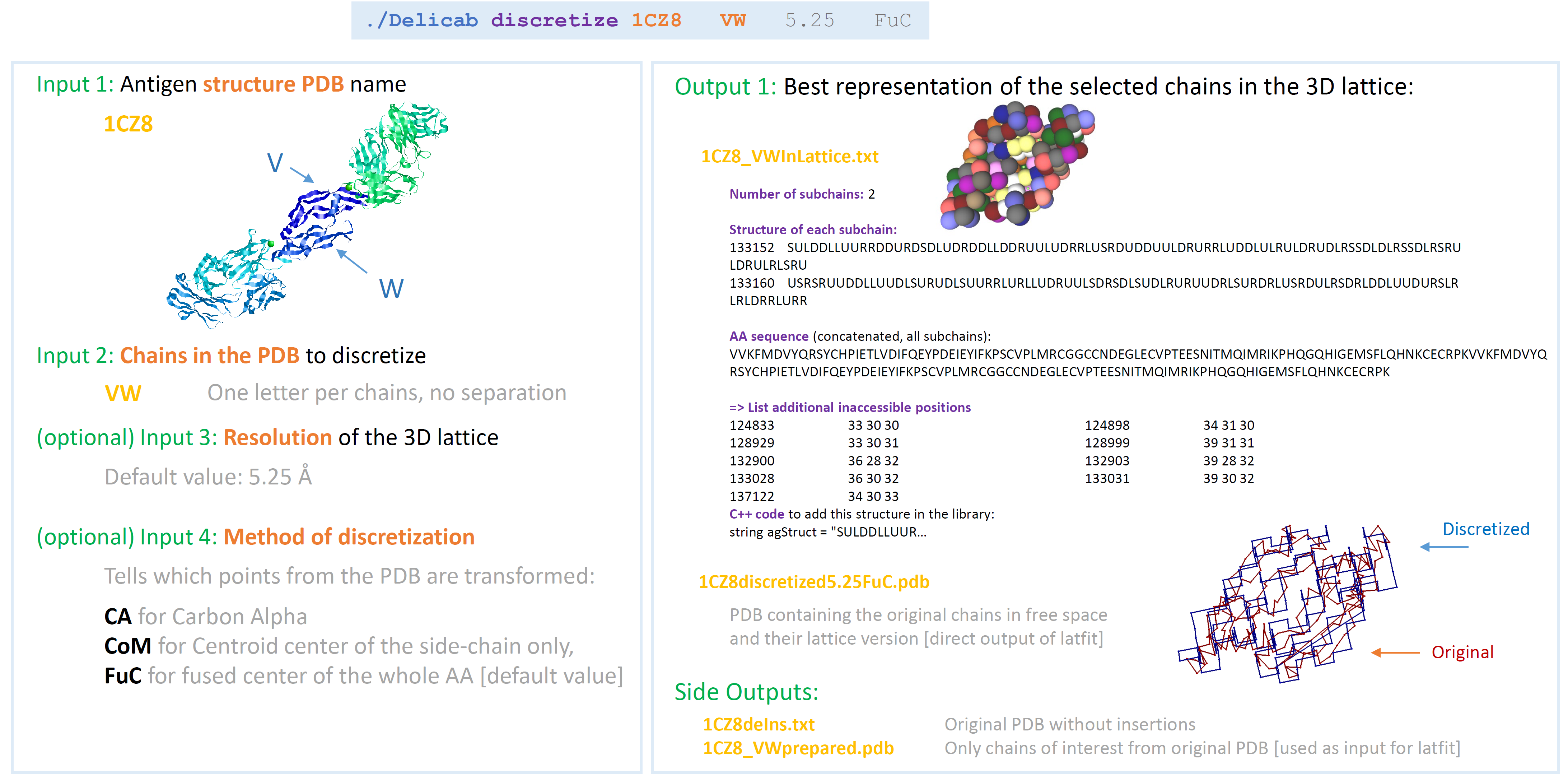
./Absolut discretize 1CZ8 VW 5.25 FuCInputs
- PDB_ID: The 4-character name of the PDB to discretize (will automatically download if not in the folder)
- Chains: The names of the chains to be discretized (one char per chain, put together)
- Resolution: The resolution of the 3D lattice [default 5.25]
- TypePos The: type of positions used for discretization [CA for Carbon Alpha, CoM for Centroid center of the side-chain only, and FuC for fused center of the whole AA – default FuC]
Outputs
- PDB and fasta files: are downloaded from the PDB server
- 1CZ8deIns.pdb: PDB with removed insertions (using pdb-tools)
- 1CZ8_VWprepared.pdb: new PDB with only the chains of interest
- 1CZ8discretized5.25FuC.pdb: discretized chains outputed from LatFit
- 1CZ8_VWInLattice.txt: Description of the discretized (lattice) antigen [Each chain is described as a starting position in the lattice (6-digits number) and a list of moves in space (straight S, up U, down D, left L, right R). See ‘info_position’ to convert lattice positions.
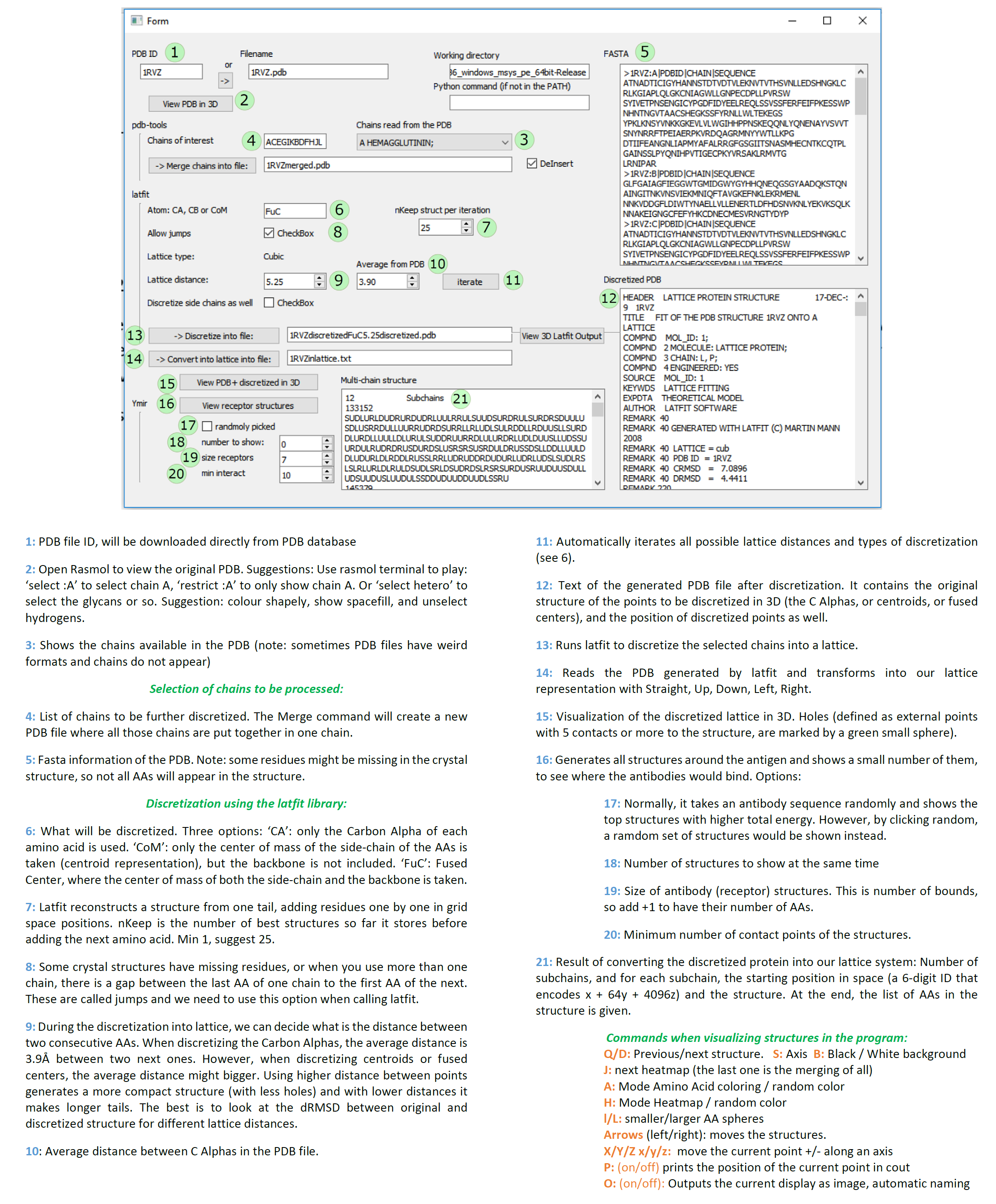
./Absolut discretizeThe inputs are decided from the user interface (and can also be provided in the command line). Identical outputs as above. Possible to export pictures during visualization inside the interface (command 'O', see documentation on visualization).
./Absolut listAntigensOutput
0 1FNS_A
1 1ADQ_A
2 1FSK_A
3 1FBI_X
4 1H0D_CThe library of lattice antigens is defined inside antigenLib.cpp. Their name is "PDB_Chains", for instance "1CZ8_VW". More than 150 lattice antigens are already available. If you wish to add your own, please manually add them into antigenLib.cpp.
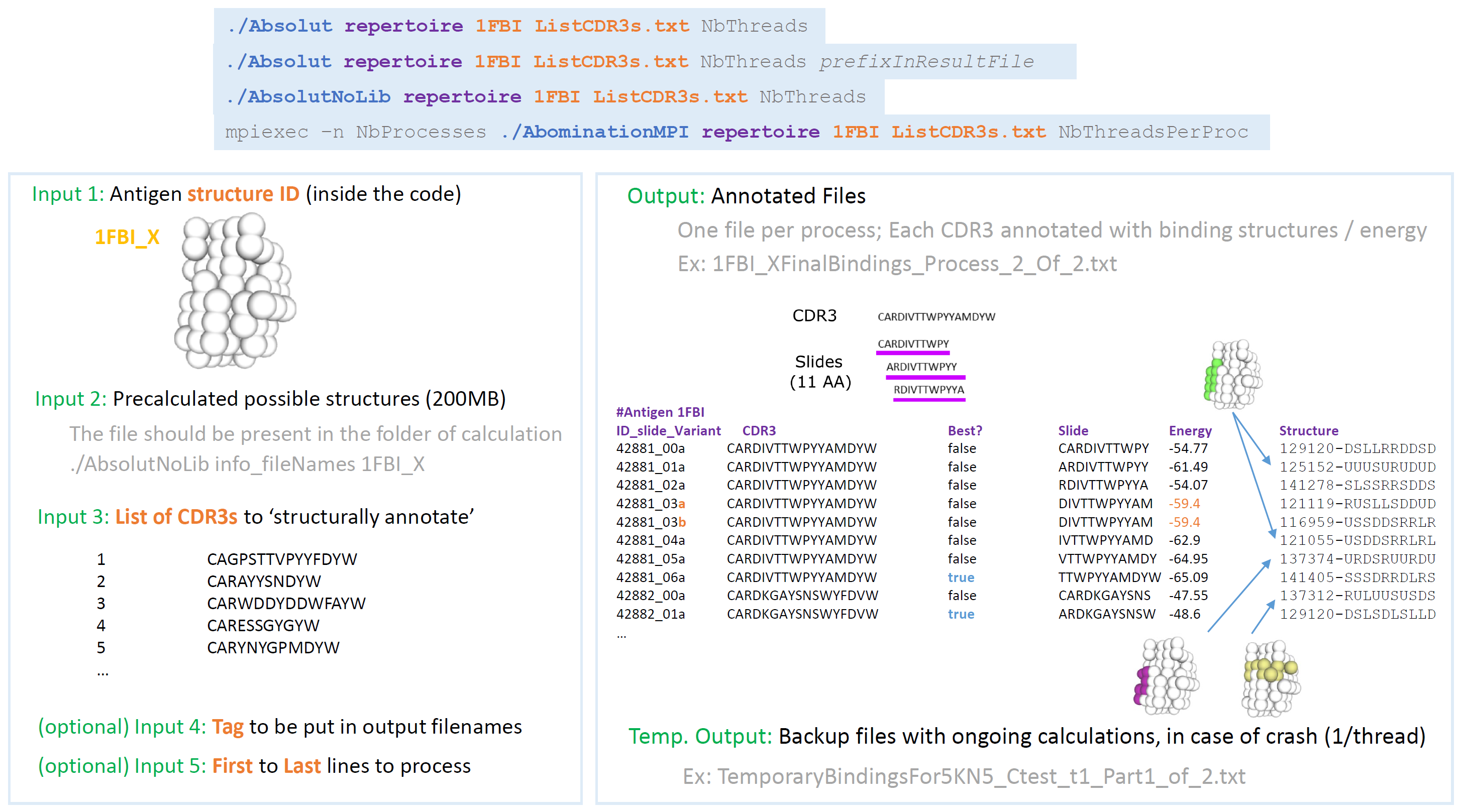
This takes a lattice antigen and a list of CDR3s sequences and returns the best binding structure of each 11-mers of each CDR3 around the antigen.
There are two ways. The singleBinding option allows to calculate for one CDR3 in particular and is good for testing. The repertoire option allows to process a text file with a list of CDR3s.
Step 1: Get requested pre-computed files (once only)
./Absolut info_fileNames 1FBI_X
wget ...Output
Pre-calculated structures are in SULSU040643e2c0a6d6343bbe8a27b079ef91-10-11-efc862c2cdef086ba79606103a3dfc62Structures.txt
use:
wget http://philippe-robert.com/Absolut/Structures/SULSU040643e2c0a6d6343bbe8a27b079ef91-10-11-efc862c2cdef086ba79606103a3dfc62Structures.txtStep 2: Calculate the bindigns, after the precomputed structure file is in the same (or parent) folder
#For one CDR3 only
./Absolut singleBinding 1FBI_X CARAAHKLARIPK#For a list of CDR3s provided in ListCDR3s.txt
./Absolut repertoire 1FBI_X ListCDR3s.txt
#Or, without MPI, the program is still paralellized to use multiple threads, for instance 20 here
./Absolut repertoire 1FBI_X ListCDR3s.txt 20
#Or, in order to use MPI, for instance using 50 threads for each MPI process
mpiexec –n NbProcesses ./AbominationMPI repertoire 1FBI_X ListCDR3s.txt 50Inputs
- Antigen_ID: The ID of the antigen in the library.
- ListCDR3s.txt: a text file with a list of CDR3s. Two columns, ID and CDR3 Amino Acid sequence, tab-separated, no header:
1 CAGPSTTVPYYFDYW
2 CARAYYSNDYW
3 CARWDDYDDWFAYW
4 CARESSGYGYW
5 CARYNYGPMDYW
6 CARGDSFDYW
7 CARVPNWDVNWSFDVW
...Note, only the CDR3s with 11 AAs or more will be considered.
- A precomputed structure file in the folder (or parent folder) where Absolut is called. This is a text file containing the list of all pre-calculated prossible bindings. You can use the wget command provided when calling ./Absolut info_fileNames. We have precalculated these files for the library antigens and made them available in http://philippe-robert.com/Absolut/Structures/ Note, it is possible to recompute the structure files for a library antigen or any new home-made antigens, by using the command singleBinding above, that will regenerate the files if not present in the folder. The repertoire option stops if the files are not available.
Outputs
- Text file called "1FBI_XFinalBindings_Process_1_Of_1.txt", containing the structural annotation on how each 11-mer (Slide) binds to the antigen, with its binding energy and its structure (position-list of moves) A new ID is generated for each slide, with the CDR3ID from the input file followed with _ and the number of the slide and a/b/c... if different structures share the best binding energy. The best way a CDR3 binds to the antigen is annotated with Best=true. This file format is called Raw Binding dataset
#Antigen 1FBI
ID_slide_Variant CDR3 Best Slide Energy Structure
42881_00a CARDIVTTWPYYAMDYW false CARDIVTTWPY -54.77 129120-DSLLRRDDSD
42881_01a CARDIVTTWPYYAMDYW false ARDIVTTWPYY -61.49 125152-UUUSURUDUD
42881_02a CARDIVTTWPYYAMDYW false RDIVTTWPYYA -54.07 141278-SLSSRRSDDS
42881_03a CARDIVTTWPYYAMDYW false DIVTTWPYYAM -59.4 121119-RUSLLSDDUD
42881_03b CARDIVTTWPYYAMDYW false DIVTTWPYYAM -59.4 116959-USSDDSRRLR
42881_04a CARDIVTTWPYYAMDYW false IVTTWPYYAMD -62.9 121055-USDDSRRLRL
42881_05a CARDIVTTWPYYAMDYW false VTTWPYYAMDY -64.95 137374-URDSRUURDU
42881_06a CARDIVTTWPYYAMDYW true TTWPYYAMDYW -65.09 141405-SSSDRRDLRS
42882_00a CARDKGAYSNSWYFDVW false CARDKGAYSNS -47.55 137312-RULUUSUSDS
42882_01a CARDKGAYSNSWYFDVW true ARDKGAYSNSW -48.6 129120-DSLSDLSLLD
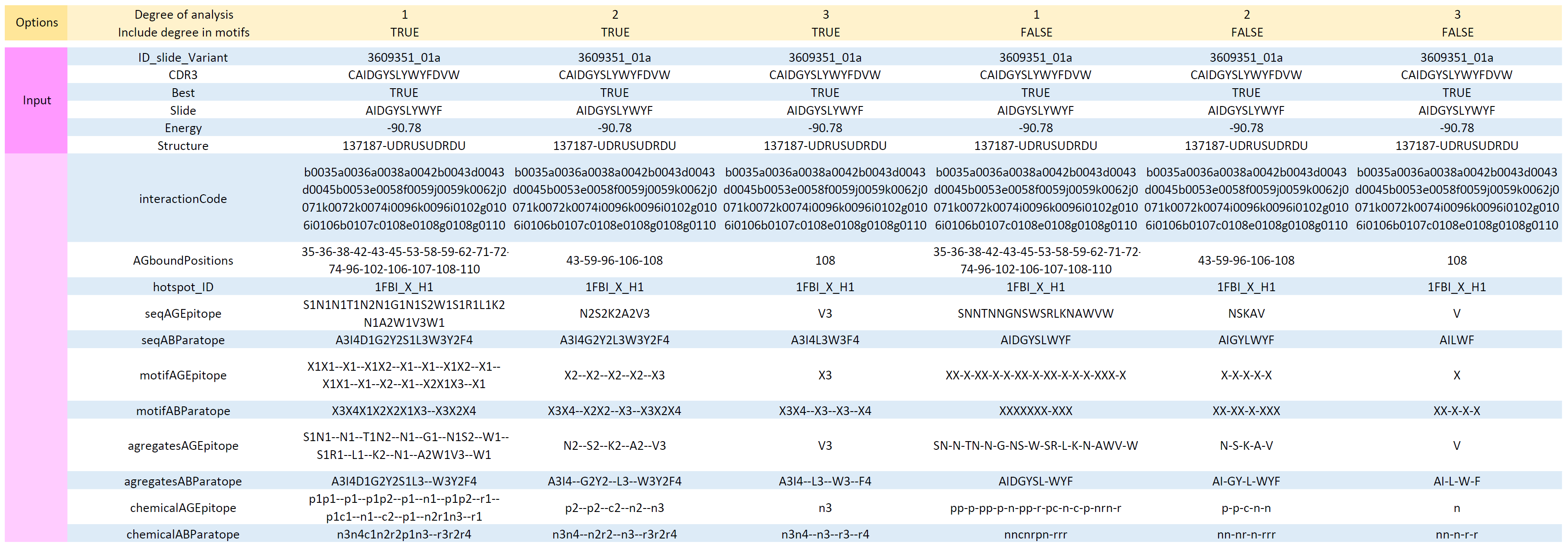
…./Absolut getFeatures 1FBI_X 1FBI_XRawBindings.txt outputFeaturesFile.txt
#or, it is possible to precise the binding degree in the analysis by adding the degree (1 here) and true/false to include the degree into the encodings
./Absolut getFeatures 1FBI_X 1FBI_XRawBindings.txt outputFeaturesFile.txt 1 trueInputs
- Antigen ID
- 1FBI_XRawBindings.txt: The raw binding file generated from the repertoire option below
- outputFeaturesFile.txt: The file that will be created with the binding features
- degree(optional): this specify that only AA interactions of degree equal or more than X (between antibody and antigen) are taken into the encoding
- showDegreeInAnalyses: this will add the degree into the coarse grained encodings, like the motif XX-X-X will become X1X2--X1--X3.
Outputs
- outputFeaturesFile.txt with the same lines as the input line, but extended with one column per feature. The features are:
Note: you might want to keep only the best Slides per CDR3 (Best=true) from the raw binding file before generating the features.
To visualize an antigen from the library
./Absolut visualize 1FBI_XTo include the bindings from a raw binding with an optional energy threshold
./Absolut visualize 1FBI_X 1FBI_XRawBindings.txt
./Absolut visualize 1FBI_X 1FBI_XRawBindings.txt -95.5./Absolut hotspots 1FBI_X 1FBI_XRawBindings.txt -95.5