MONAI Segment is a mercure module for rapid deployment of segmentation models hosted in the MONAI model zoo.
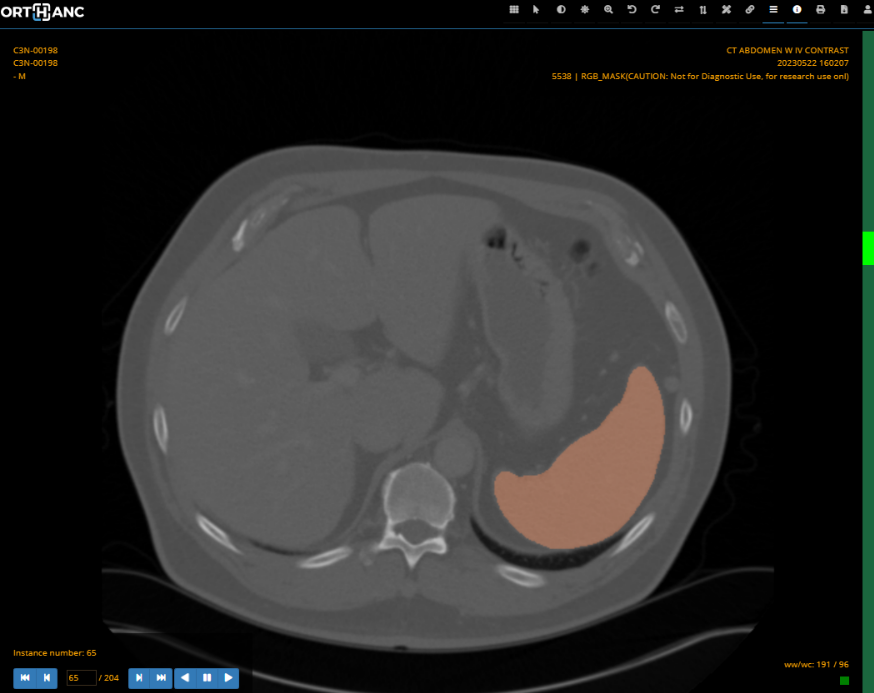
This module is available as a docker image that can be added to an existing mercure installation using docker tag : mercureimaging/mercure-monaisegment. It will perform segmentation of the spleen in CT images using the Spleen ct segmentation MONAI bundle. The code was adapted from the MONAI Deploy bundle app tutorial.
The code can be simply modified to rapidly deploy other segmentation models in the MONAI model zoo.
Follow instructions on mercure website on how to add a new module. Use the docker tag mercureimaging/mercure-monaisegment.
Install virtual box and vagrant and follow jupyter notebook tutorial tutorial_mercure-MonaiSegment.ipynb (less than 1hr to complete).
- Clone repo.
- Build Docker container locally by running make (modify makefile with new docker tag as needed).
- Test container :
docker run -it -v /input_data:/input -v /output_data:/output --env MERCURE_IN_DIR=/input --env MERCURE_OUT_DIR=/output *docker-tag*
Segmentations are written to specified output directory in the following formats :
- DICOM SEG
- DICOM RGB masks (segmentations overlaid on background images)
- Default model 'spleen_ct_segmentation' bundle included in Dockerfile settings.
- To select a different model, set MONAI_BUNDLE_URL to MONAI Model Zoo bundle link address.
- Current requirements:
- MONAI bundle must support torchscript i.e. include model.ts file
- MONAI bundle must conform to spec.
- MONAI bundle must contain a segmentation model
- May require further preprocessing or transform operators prior to inference to function correctly.