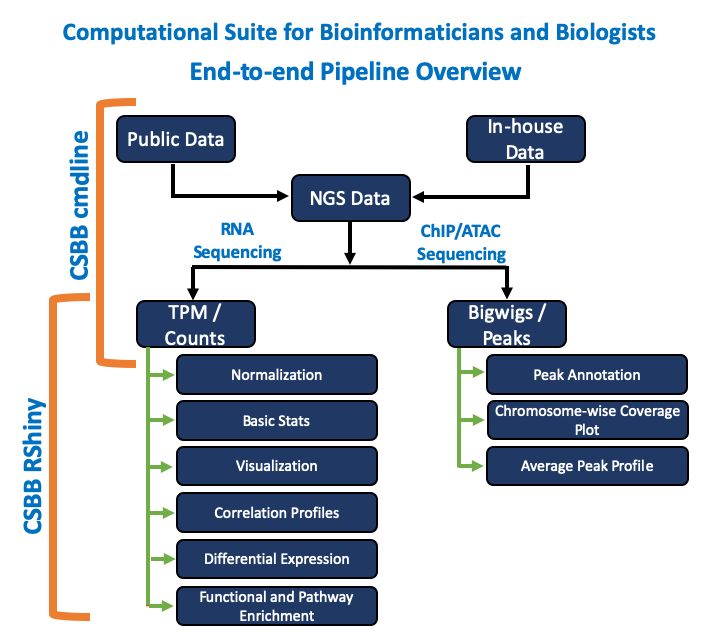
Computational Suite for Bioinformaticians and Biologists (CSBB), is a RShiny application developed with an intention to empower researchers from wet and dry lab to perform downstream Bioinformatics analysis. CSBB powered by RShiny is packed with 7 modules Visualization, Normalization, Basic Stats, Differential Expression, Correlation Profiles, Function/Pathway Enrichment and ChIP-ATAC Seq Analysis. These modules are designed in order to help researchers design a hypothesis or answer research questions with little or no expertise in Bioinformatics. CSBB is also available as a command line application and has Next generation sequencing data processing capabilities. New modules and functionalities will be added periodically
library(shiny) library(servr) library(ggplot2) library(pheatmap) library(M3C) library(RUVSeq) library(scales) library(dtwclust) library(dplyr) library(ggcorrplot) library(tibble) library(ReactomePA) library(org.Hs.eg.db) library(org.Mm.eg.db) library(AnnotationDbi) library(EnhancedVolcano) library(ChIPseeker) library(TxDb.Hsapiens.UCSC.hg19.knownGene) library(TxDb.Mmusculus.UCSC.mm10.knownGene) library(TxDb.Hsapiens.UCSC.hg38.knownGene) library(TxDb.Mmusculus.UCSC.mm9.knownGene) library(clusterProfiler)