The goal of this repository is to provide R users an example of how to automate simulation studies on high-performance computing platforms (a.k.a. the cluster). The pipeline was initially written for the Slurm scheduling system, but can be easily modified for other systems.
The main idea of this pipeline is to use an R script to compose jobs via
system call, where each job corresponds to a batch of replications under
the same simulation setting. These replications are implemented via the
Slurm job array feature using the flag
--array.
We use a simulation study of the central limit theorem (CLT) to demonstrate the pipeline. In this example, we simulate random samples (X1, …, Xn) of size n ∈ {5, 10, 50, 100, 500, 1000, 5000, 10000} from independent and identical Poisson distribution with mean λ = 3. We calculate the sample mean X̄ of the n random sample. This process is repeated for 100000 times to generate an empirical distribution of X̄. We hypothesize this empirical distribution should closely match to a normal distribution of mean 3 and variance 3/n, particularly when n grows large. To examine our hypothesis, we record the mean, median, variance of the empirical distribution. This concludes as one replication, and we have nit = 1000 replications for each setting of n.
To put the simulation study in the context of the pipeline, we have
- one simulation parameter: the size of random samples n
- the data generating process: 100000 samples of (X1, …, Xn) where Xi ∼ Poisson(λ = 3) i.i.d. for i = 1, …, n
- the model and its output: the empirical distribution of X̄ and its mean, median, variance
- the number of replications for each simulation setting: nit = 1000
-
Copy three code files (including
start_sim.R,slurm_job_config.job,main.R) in the folderCode-copy_meto your working directory on the cluster. -
Replace the code section in the
main.Rfile with your simulation code -
Replace the directory paths with your preferred path
- Global
search
the phrase
TODO: Replace path herein the code files - We recommend to use absolute path instead of relative path in your files
- Global
search
the phrase
-
In the log-in node of the cluster, source the file
start_sim.Rin an R console or runR CMD BATCH start_sim.Rin the command line within your working directory, i.e. the directory that contains the three code files- You still need to load the R module before running the R script in your preferred way.
-
Wait to see the simulation output in the designated storage folder/directory
- If there was something wrong with the simulation, go to your log folder/directory
We provide simple practices to help you familiarize with the pipeline, please consider to improve the current simulation study by
- examining more simulation parameters, e.g. more options of the sample distribution Xn
- replicating the simulation study for nit ∈ {100, 10000} times per each simulation setting
- summarizing the empirical distribution of X̄ differently, e.g. kurtosis, skewness, or normality test
The pipeline is built upon the premise that running each repetition via different jobs (i.e. 1000 jobs where each job requires a single core on a node) is more efficient than running all repetitions within a giant loop that has some parallel capacity (i.e. 1 job that requires 1 node with 44 CPUs and 2 days of computation time). This is generally true when you can create more jobs (without long queuing time) than the number of cores that available for use. In addition, it takes a lot longer time to request a single node with a great number of CPUs.
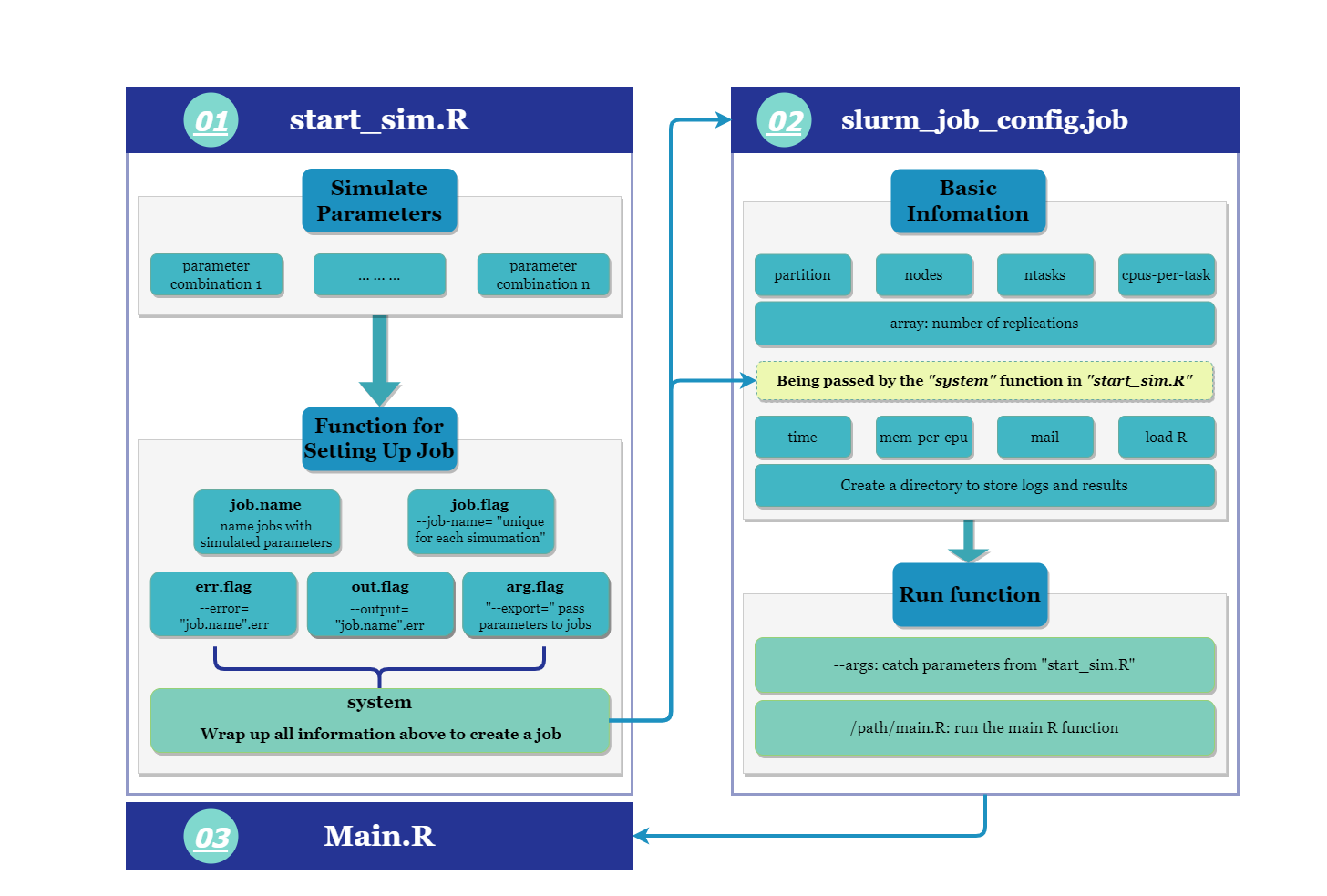
The main idea of the pipeline is to create an R interface sim_start.R
where you can set up parameters of the simulation study. Then, the R
interface will help to create job arrays via sys.call. Each simulation
setting has a job array that contains independent replications of
preferred size. The parameters that are necessary to create a simulation
setting is passed to the job scheduling script slurm_job_config.job
and further to the R code main.R that contains the data generating
process. In other words, we can use the same job script and data
generating process with different sets of simulation parameters.
| Process of Setting up Cluster Jobs using R |
|---|
 |
Please feel free to ask us questions or report bugs via Issues tab of the repository. We advise you to search if a similar question or bug was previously issued before filing your own. You can search and filter by using the search box or the labels. Two templates are provided to report, where you need to describe your problem and provide a minimal reproducible example.
-
Modularize your code
When you write your simulation code, try to think how to best modularize your code sections. Each module can be used at multiple locations/simulation settings without writing them repetitively. In addition, Modularization provides addition convinience during the debugging process. Instead of going through a long script, you can test out each module to locate the bug. To start with, you can pack you code into functions, where each function have a dedicated purpose. For example, you can write a function for data generating process, a function for model training, and a function for model testing.
-
Test out your code before deploying on the cluster
It is hardly possible that anyone can set up their simulation study correctly the first time. Hence, it is good to test the code before deployment on the cluster. It will save yoursolve from reading weired error message and cleaning up log files. The best practice is to start small, for example run one of the simulation settings with small number of repetitions. If it works, proceed to scale up the simulation study.
-
Choose the correct cluster partition
_ Coosing the cluster partition has a direct effect on how many jobs you can run simultaneous, how long it takes to start a job, and hence decides how fast you can get the study results. It requires optimization based on your own needs if you prefer to getting your result back as soon as possible. The shorter time you request, the more job you can run parallel. However, if you didn’t have enough computation time for your job, you would simply lose the efforts you had._
-
Save simulated data Vs Save simulation results
These are the two strategies for the whole simulation study (data generation + analysis with models). Each has its own merit. Saving simulated data gives you flexibility to conduct granular investigation, say each replication, which is helpful when debugging or thoroughly understand the behavour of models. However, it is more laborious to finish the whole simulation study, as another process to conduct the acutal analysis is required. In addition, it requires more storage room for the generated data. In contrast, having on integrated process that contains both data generation and analysis requires less effort to set up and manage. However, it loses some of the transparency to investigate case-by-case. (Not impossible, use the saved seed number/iteration number)
- Another tool to integrate your simulation study systematically is
the R workflow package
targets. It provides support for high-performance computing and cloud integration. However, it takes some effort to set up, particularly if you don’t have admin role of the system or great IT support.
- Morris, White, and Crowther (2019) developed ADEMP framework for planning simulation studies. The authors gave concrete examples and guidelines on what to consider in each step of the planning.
Morris, Tim P, Ian R White, and Michael J Crowther. 2019. “Using simulation studies to evaluate statistical methods.” Statistics in Medicine, January. https://doi.org/10.1002/sim.8086.