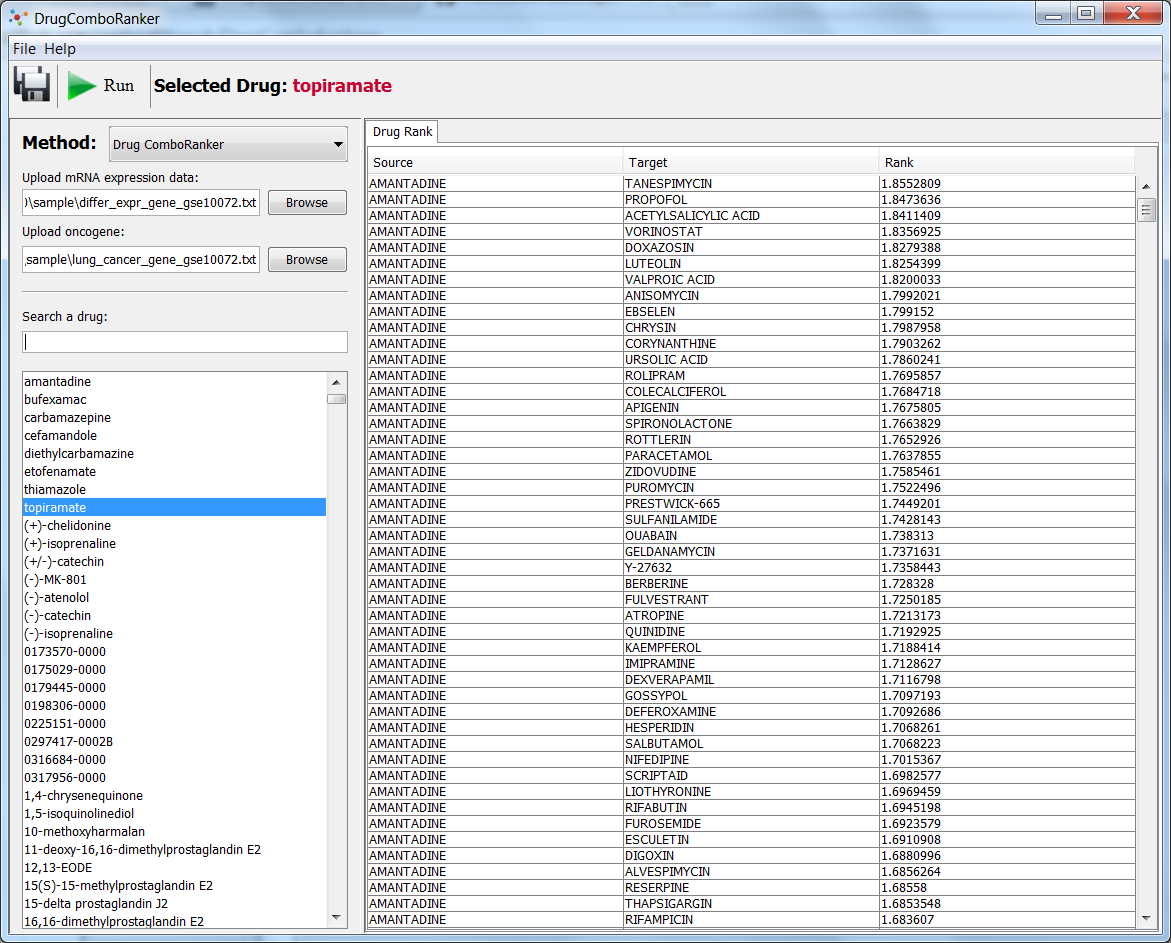
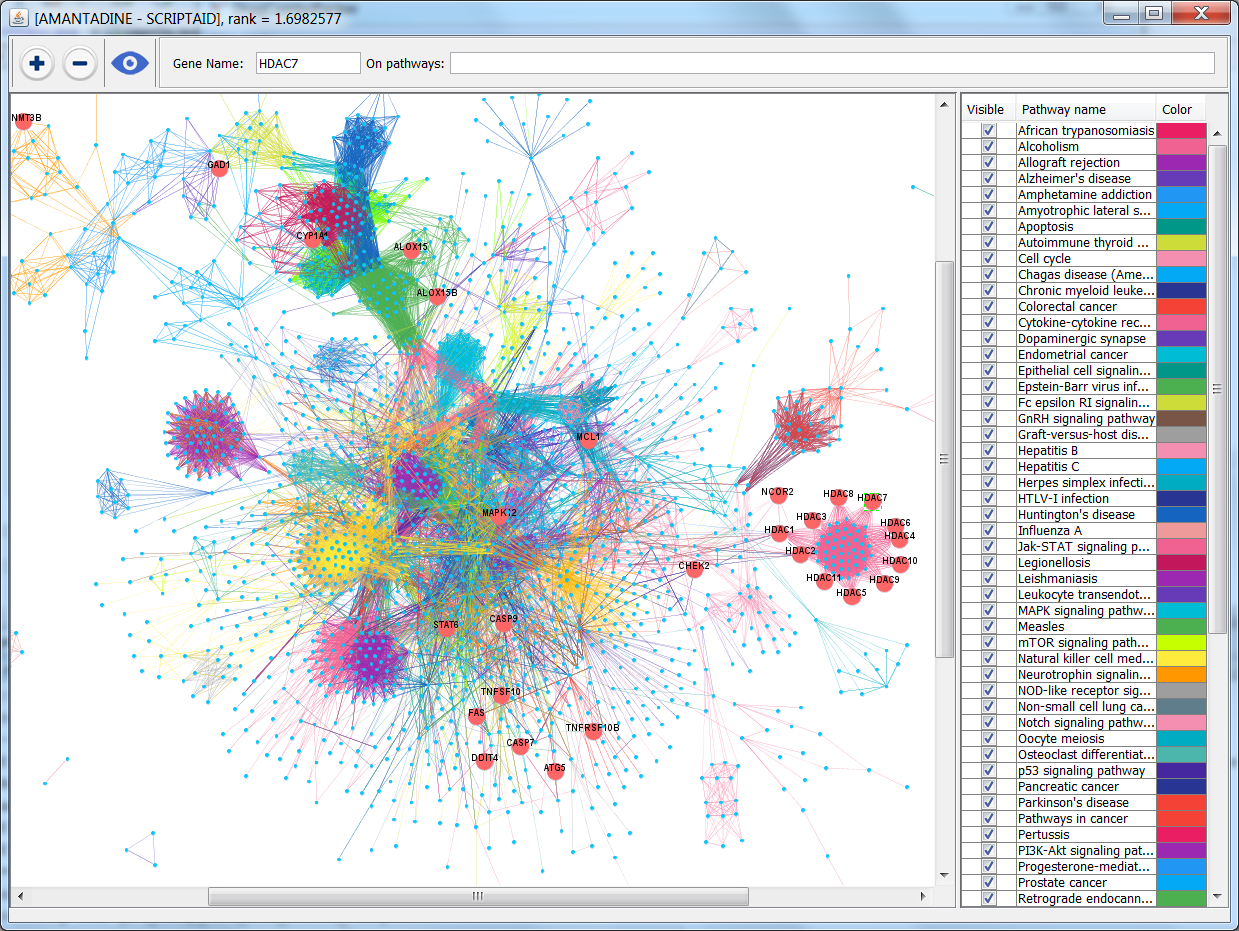
DrugComboRanker is a computational tool that prioritizes synergistic drug combinations and uncovers their mechanisms of action. It can identify drug combinations by searching for drugs whose targets are enriched in the complementary signaling modules of the disease signaling network to predict top ranked drug combinations and map the drug targets on the disease signaling network to highlight the mechanisms of action of the drug combinations.
There are no curative anticancer drugs, and drug resistance is often acquired after drug treatment. One of the reasons is that cancers are complex diseases, regulated by multiple signaling pathways and cross talks among the pathways. Drug combinations should reduce drug resistance and improve patients’ outcomes. In clinical practice, the ideal and feasible drug combinations are combinations of existing Food and Drug Administration-approved drugs or bioactive compounds that are already used on patients or have entered clinical trials and passed safety tests. These drug combinations could directly be used on patients with less concern of toxic effects. However, there is no effective computational approach to search effective drug combinations from the enormous number of possibilities.
In this study, we developed a novel systematic computational tool, DrugComboRanker, to prioritize synergistic drug combinations and uncover their mechanisms of action. We first built a drug functional network based on their genomic profiles, and partitioned the network into numerous drug network communities using a Bayesian non-negative matrix factorization approach. As drugs within an overlapping community share common mechanisms of action, we next uncover potential targets of drugs by applying a recommendation system on drug communities. We meanwhile build disease-specific signaling networks based on patients’ genomic profiles and interactome data. We then identify drug combinations by searching drugs whose targets are enriched in the complementary signaling modules of the disease signaling network. The novel method was evaluated on lung adenocarcinoma and endocrine receptor positive breast cancer, then compared with other drug combination approaches. These case studies discovered a set of effective drug combinations top ranked in our prediction list and mapped the drug targets on the disease signaling network to highlight the mechanisms of action of the drug combinations.
DrugComboRanker: drug combination discovery based on target network analysis. Lei Huang, Fuhai Li, Jianting Sheng, Xiaofeng Xia, Jinwen Ma, Ming Zhan and Stephen T.C. Wong, 2014. 30:i228–i236
http://70.35.195.67/drugcomboranker/
https://github.com/methodistsmab/DrugComboRanker/blob/master/DrugComboRanker_user_guide_v1.pdf
https://github.com/methodistsmab/DrugComboExplorer/blob/master/End_User_License_Agreement.pdf
Lei Huang
Email: lhuang@houstonmethodist.org
Xiaohui Yu
Email: xyu2@houstonmethodist.org
Wong, Stephen (PI)
Email: STWong@houstonmethodist.org