This repository contains the examples used in the lecture and some general tips for making more reproducible bioinformatic research.
As an example of the concepts introduced in the lecture the repository contains a simple pipeline. The pipeline is designed to be ran in a docker container.
The pipeline is intended to illustrate some programming concepts rather than conducting any meaningful analysis, hence the overenginered design and simple function.
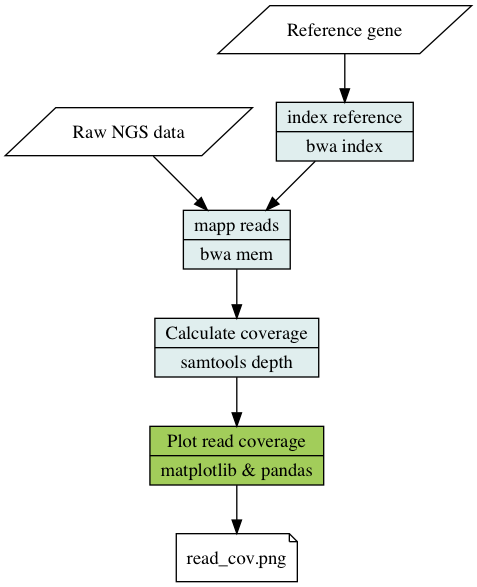
The pipeline aligns reads, using BWA, from a NGS sample to a reference sequence in fasta format. The number of reads covering a each base in the reference sequence is calculated using samtools depth and plotted using matplotlib and pandas.
- Docker Engine
- https://docs.docker.com/engine/installation/ ( Version 17.09.0+ )
- GNU wget
- for downloading data with download_example_files.sh ( Version 1.19.2+ )
- Launch the Docker.app that you’ve installed and provide the credentials as needed for the initial setup.
- In the Docker.app preferences, make the following adjustments.
- Advanced: Dedicate >2 CPUs and >4GB of RAM to Docker.
- Apply and Restart
To download the pipeline you need to first clone the repository to your computer with the following command.
git clone https://mhkj@bitbucket.org/mhkj/reproducible-bioinformatic-research.gitThe pipeline comes with a script for downloading example files to the folder ./volumes/data. The example is a clinical isolate from a Staphylococcus aureus MRSA outbreak at a special care baby unit (SCBU) in Cambridge [fn:1].
./download_example_files.shTo build, run and stop the pipeline you need to do the following steps. You can to them by either running the following shell commands in you terminal or using the Makefile with the arguments build, run and stop. The makefile runs the same commands, it’s just a convenient wrapper.
To run the pipeline, you need to build a container image using either the following shell command, you only need to do this once since the build image is then stored on your computer.
docker build . -t example-pipelineAfter the image has been build you need to boot an instance of the image with the following command. This will take you into the command line of the booted container. NOTE Do not use sudo inside a container since it might create files, or give access to files, outside of the container and thus break the isolated environment.
docker run \
--name=example-pipeline \
-it \
--volume $PWD/volumes/temp_files:/tmp/example_pipeline \
--volume $PWD/volumes/results:/usr/src/example_pipeline/results \
--volume $PWD/volumes/data:/usr/src/example_pipeline/data \
example-pipelineUse the following command to run the pipeline inside the booted container.
./run_pipeline.py --threads 2 \
--fasta-file ./data/blaz.fna \
--fastq-file ./data/err418327_1.fastq.gz \
--fastq-file ./data/err418327_2.fastq.gz \
results/read_coverage.pngWhen you have finished running the sample. Exit the container with ctrl-d. The resulting plot should now be located in ./volumes/results/ and the generated alignments and intermediary files should be located in ./volumes/temp_files. If you want to run additional samples in the pipeline, please empty the folders but dont remove them since it could cause errors with file permissions in the docker container.
When you are finished. Stop all running containers with the following command following command.
docker container stop example-pipeline
docker container rm example-pipeline[fn:1] Simon R Harris, Edward JP Cartwright, M Estée Török, Matthew TG Holden, Nicholas M Brown, Amanda L Ogilvy-Stuart, Matthew J Ellington, Michael A Quail, Stephen D Bentley, Julian Parkhill, Sharon J Peacock, Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study, In The Lancet Infectious Diseases, Volume 13, Issue 2, 2013, Pages 130-136, ISSN 1473-3099, https://doi.org/10.1016/S1473-3099(12)70268-2.