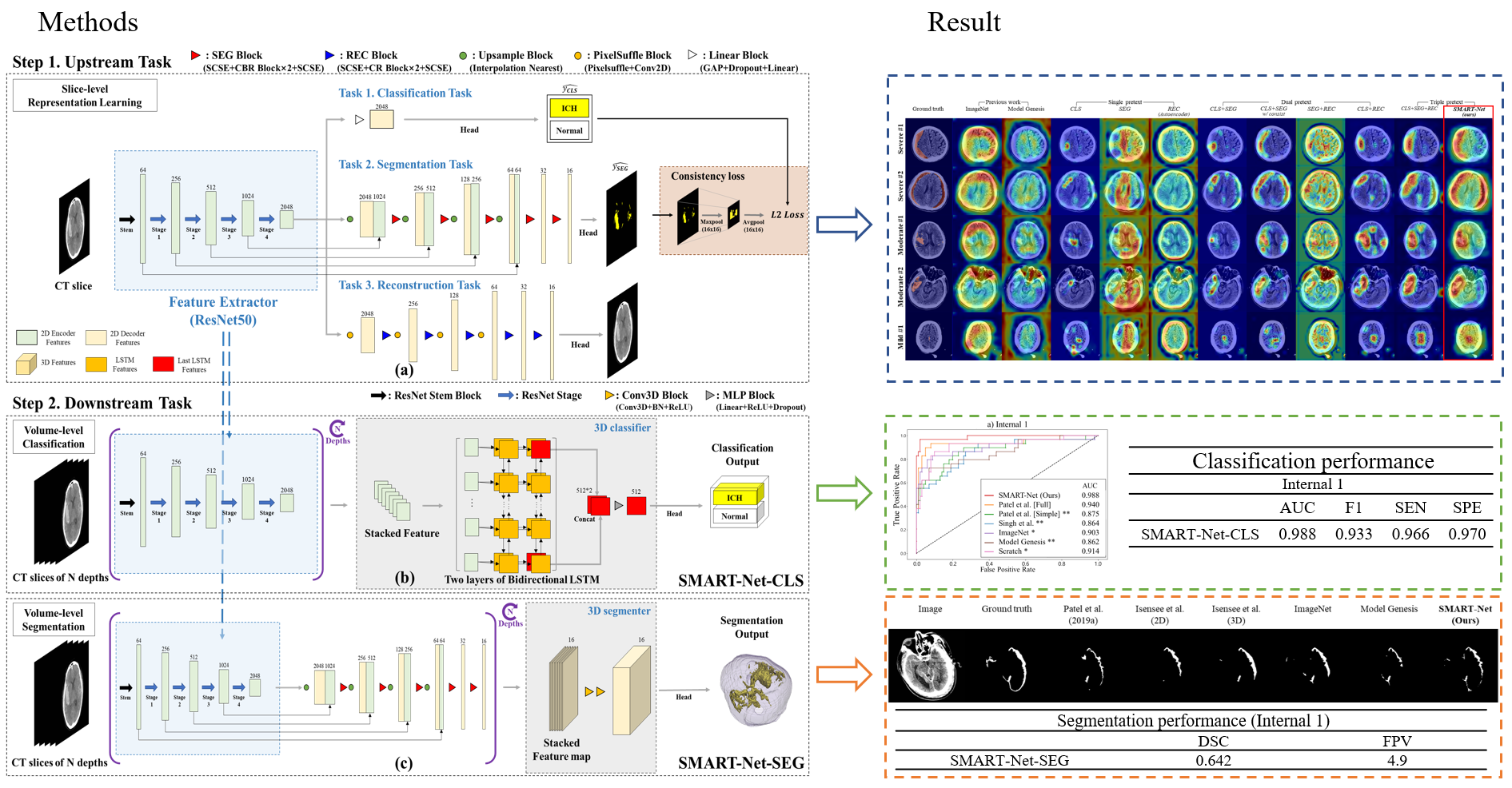
We proposed a supervised multi-task aiding representation transfer learning network called SMART-Net.
- Develop a robust feature extractor of brain hemorrhage in head & neck NCCT through three kinds of multi-task representation learning.
- Propose the consistency loss to alleviate the disparity of two pretext tasks' heads, resulting in improved transferability and representation. Connect the feature extractor with the target-specific 3D operator via transfer learning to expand volume-level tasks.
- Explore relationships of the proposed multi-pretext task combinations and perform ablation studies on optimal 3D operators for volume-level ICH tasks.
- Validate the model on multiple datasets with previous methods and ablation studies for the robustness and practicality of our method.
This repository provides the official implementation of training SMART-Net as well as the usage of the pre-trained SMART-Net in the following paper:
Improved performance and robustness of multi-task representation learning with consistency loss between pretexts for intracranial hemorrhage identification in head CT
Sunggu Kyung1, Keewon Shin, Hyunsu Jeong, Ki Duk Kim, Jooyoung Park, Kyungjin Cho, Jeong Hyun Lee, Gil-Sun Hong, and Namkug Kim
MI2RL LAB
(Under revision...) Medical Image Analysis (MedIA)
- Linux
- Python 3.8.5
- PyTorch 1.8.0
$ git clone https://github.com/babbu3682/SMART-Net.git
$ cd SMART-Net/
$ pip install -r requirements.txtFor your convenience, we have provided few 3D nii samples from Physionet publish dataset as well as their mask labels.
You can use your own data using the dicom2nifti for converting from dicom to nii.
- The processed hemorrhage directory structure
datasets/samples/
train
|-- sample1_hemo_img.nii.gz
|-- sample1_hemo_mask.nii
|-- sample2_normal_img.nii.gz
|-- sample2_normal_mask.nii
.
.
.
valid
|-- sample9_hemo_img.nii.gz
|-- sample9_hemo_mask.nii
|-- sample10_normal_img.nii.gz
|-- sample10_normal_mask.nii
.
.
.
test
|-- sample20_hemo_img.nii.gz
|-- sample20_hemo_mask.nii
|-- sample21_normal_img.nii.gz
|-- sample21_normal_mask.nii
.
.
.
- Up_SMART_Net
- Up_SMART_Net_Dual_CLS_SEG
- Up_SMART_Net_Dual_CLS_REC
- Up_SMART_Net_Dual_SEG_REC
- Up_SMART_Net_Single_CLS
- Up_SMART_Net_Single_SEG
- Up_SMART_Net_Single_REC
+ train: We conducted upstream training with three multi-task including classificatiom, segmentation and reconstruction.
python train.py \
--data-folder-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/datasets/samples' \
--model-name 'Up_SMART_Net' \
--batch-size 10 \
--epochs 1000 \
--num-workers 4 \
--pin-mem \
--training-stream 'Upstream' \
--multi-gpu-mode 'DataParallel' \
--cuda-visible-devices '2, 3' \
--gradual-unfreeze 'True' \
--print-freq 1 \
--output-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/up_test'+ test (slice-wise for slice-level): We conducted upstream training with three multi-task including classificatiom, segmentation and reconstruction.
python test.py \
--data-folder-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/datasets/samples' \
--test-dataset-name 'Custom' \
--slice-wise-manner "True" \
--model-name 'Up_SMART_Net' \
--num-workers 4 \
--pin-mem \
--training-stream 'Upstream' \
--multi-gpu-mode 'Single' \
--cuda-visible-devices '2' \
--print-freq 1 \
--output-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/up_test' \
--resume '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/up_test/epoch_0_checkpoint.pth'+ test (stacking slice for volume-level): We conducted upstream training with three multi-task including classificatiom, segmentation and reconstruction.
python test.py \
--data-folder-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/datasets/samples' \
--test-dataset-name 'Custom' \
--slice-wise-manner "False" \
--model-name 'Up_SMART_Net' \
--num-workers 4 \
--pin-mem \
--training-stream 'Upstream' \
--multi-gpu-mode 'Single' \
--cuda-visible-devices '2' \
--print-freq 1 \
--output-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/up_test' \
--resume '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/up_test/epoch_0_checkpoint.pth'- Down_SMART_Net_CLS
- Down_SMART_Net_SEG
+ train: We conducted downstream training using multi-task representation.
python train.py \
--data-folder-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/datasets/samples' \
--model-name 'Down_SMART_Net_CLS' \
--batch-size 2 \
--epochs 1000 \
--num-workers 4 \
--pin-mem \
--training-stream 'Downstream' \
--multi-gpu-mode 'DataParallel' \
--cuda-visible-devices '2, 3' \
--gradual-unfreeze 'True' \
--print-freq 1 \
--output-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/down_cls_test' \
--from-pretrained '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/[UpTASK]ResNet50_ImageNet.pth' \
--load-weight-type 'encoder'+ test: We conducted upstream training with three multi-task including classificatiom, segmentation and reconstruction.
python test.py \
--data-folder-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/datasets/samples' \
--test-dataset-name 'Custom' \
--slice-wise-manner 'False' \
--model-name 'Down_SMART_Net_CLS' \
--num-workers 4 \
--pin-mem \
--training-stream 'Downstream' \
--multi-gpu-mode 'Single' \
--cuda-visible-devices '2' \
--print-freq 1 \
--output-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/down_cls_test' \
--resume '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/down_cls_test/epoch_0_checkpoint.pth'
+ train: We conducted downstream training using multi-task representation.
python train.py \
--data-folder-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/datasets/samples' \
--model-name 'Down_SMART_Net_SEG' \
--batch-size 2 \
--epochs 1000 \
--num-workers 4 \
--pin-mem \
--training-stream 'Downstream' \
--multi-gpu-mode 'DataParallel' \
--cuda-visible-devices '2, 3' \
--gradual-unfreeze 'True' \
--print-freq 1 \
--output-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/down_seg_test' \
--from-pretrained '/workspace/sunggu/1.Hemorrhage/SMART-Net/up_test/epoch_0_checkpoint.pth' \
--load-weight-type 'encoder'+ test: We conducted upstream training with three multi-task including classificatiom, segmentation and reconstruction.
python test.py \
--data-folder-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/datasets/samples' \
--test-dataset-name 'Custom' \
--slice-wise-manner 'False' \
--model-name 'Down_SMART_Net_SEG' \
--num-workers 4 \
--pin-mem \
--training-stream 'Downstream' \
--multi-gpu-mode 'Single' \
--cuda-visible-devices '2' \
--print-freq 1 \
--output-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/down_seg_test' \
--resume '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/down_seg_test/epoch_0_checkpoint.pth'
- Up_SMART_Net
- Down_SMART_Net_CLS
- Down_SMART_Net_SEG
+ inference: We conducted downstream training using multi-task representation.
python inference.py \
--data-folder-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/datasets/samples' \
--test-dataset-name 'Custom' \
--slice-wise-manner "True" \
--model-name 'Up_SMART_Net' \
--num-workers 4 \
--pin-mem \
--training-stream 'Upstream' \
--multi-gpu-mode 'Single' \
--cuda-visible-devices '2' \
--print-freq 1 \
--output-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/up_test' \
--resume '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/up_test/epoch_0_checkpoint.pth'
+ inference: We conducted downstream training using multi-task representation.
python inference.py \
--data-folder-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/datasets/samples' \
--test-dataset-name 'Custom' \
--slice-wise-manner "False" \
--model-name 'Down_SMART_Net_CLS' \
--num-workers 4 \
--pin-mem \
--training-stream 'Downstream' \
--multi-gpu-mode 'Single' \
--cuda-visible-devices '2' \
--print-freq 1 \
--output-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/down_cls_test' \
--resume '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/down_cls_test/epoch_0_checkpoint.pth'
+ inference: We conducted downstream training using multi-task representation.
python inference.py \
--data-folder-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/datasets/samples' \
--test-dataset-name 'Custom' \
--slice-wise-manner "False" \
--model-name 'Down_SMART_Net_SEG' \
--num-workers 4 \
--pin-mem \
--training-stream 'Downstream' \
--multi-gpu-mode 'Single' \
--cuda-visible-devices '2' \
--print-freq 1 \
--output-dir '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/down_seg_test/pred_nii' \
--resume '/workspace/sunggu/1.Hemorrhage/SMART-Net/checkpoints/down_seg_test/epoch_0_checkpoint.pth'
⏳ It's scheduled to be uploaded soon.
⏳ It's scheduled to be uploaded soon.
For personal information security reasons of medical data in Korea, our data cannot be disclosed.
If you use this code for your research, please cite our papers:
⏳ It's scheduled to be uploaded soon.
We build SMART-Net framework by referring to the released code at qubvel/segmentation_models.pytorch and Project-MONAI/MONAI. This is a patent-pending technology.
Project is distributed under MIT License