A Python tool to extract and align genes, introns, and intergenic spacers across hundreds of plastid genomes using associative arrays
The multiple sequence alignment (MSA) of a set of plastid genomes is challenging. At least five factors are responsible for this challenge:
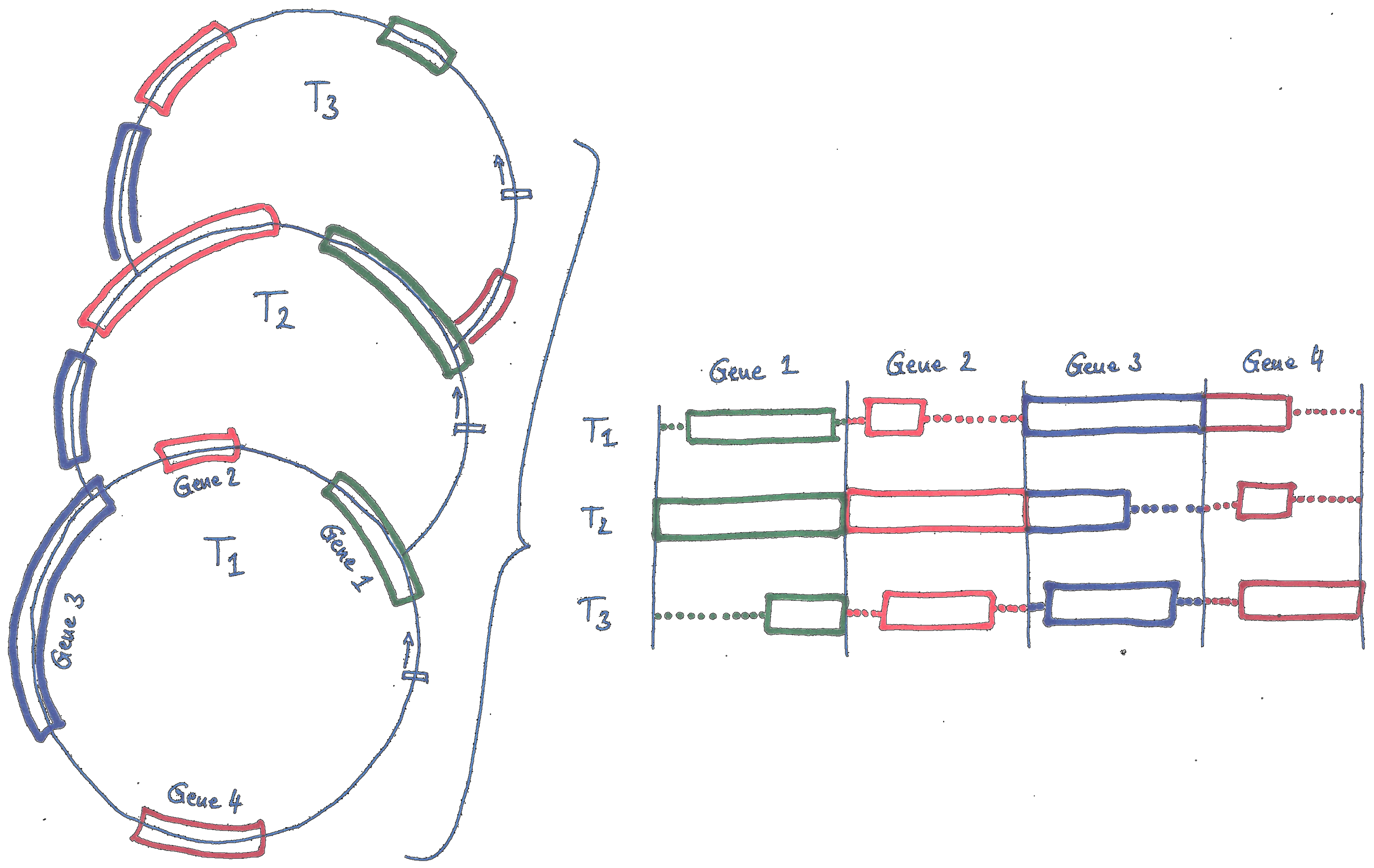
- First, the plastid genome is a mosaic of individual genome regions. A MSA procedure must identify, extract, group, and align homologous regions across genomes.
- Second, many plastid genomes exhibit sequence annotation errors regarding gene position and/or gene name. A MSA procedure must automatically remove incorrectly annotated regions from the alignment procedure.
- Third, plastid genomes comprise both coding and noncoding genome regions, which differ in their optimal alignment strategy (i.e., amino acid-based alignment for genes, nucleotide-based alignment for introns and intergenic spacers). A MSA procedure must automatically employ the best-fitting alignment strategy.
- Fourth, contemporary plastid genome investigations comprise hundreds, if not thousands, of complete plastid genomes. A MSA procedure must perform sequence alignment within reasonable time frames (i.e., hours instead of days).
- Fifth, any user-specified exclusion of a genome region from the alignment would be prohibitively complex after the alignment step. A MSA procedure must facilitate the automatic removal of user-specified genome regions.
The software plastburstalign accommodates these and more challenges: it constitutes a MSA procedure that extracts and aligns genes, introns, and intergenic spacers across hundreds or thousands of input plastid genomes.
- Extraction of all genome regions representing one of three different marker types (i.e., genes, introns, or intergenic spacers) from set of input plastid genomes, followed by grouping and alignment of the extracted regions
- Automatic exon splicing:
- automatic merging of all exons of any cis-spliced gene
- automatic grouping of all exons of any trans-spliced gene (e.g., rps12), followed by merging of adjacent exons [see
ExonSpliceHandlerfor both]
- Automatic quality control to evaluate if extracted genes are complete (i.e., valid start and stop codon present)
- Automatic removal of
- any duplicate regions (i.e., relevant for regions duplicated through the IRs)
- regions that do not fulfill a minimum, user-specified sequence length
- regions that do not fulfill a minimum, user-specified number of taxa of the dataset that the region must be found in [see
DataCleaningfor both] - any user-specified genome region (i.e., gene, intron, or intergenic spacer)
- Automatic determination if DNA sequence alignment based on amino acid (for genes) or nucleotide (for introns and intergenic spacers) sequence information
- Rapid DNA sequence extraction and alignment due to process parallelization using multiple CPUs [see
_nuc_MSA()]
- Automatic concatenation of genome regions either in alphabetic order or based on location in genome (first input genome used as reference)
- Automatic standardization of tRNA gene names to accommodate letter case differences among the gene annotations of different input genomes (e.g., for anticodon and amino acid abbreviations of tRNAs) [see
clean_gene()] - Simple installation due to automatic retrieval of third-party alignment software (MAFFT)
- Production of informative logs; two detail levels:
- default (suitable for regular software execution)
- verbose (suitable for debugging)
- Provisioning of explanation if and why a genome region could not be extracted from an input genome
- Set of complete plastid genomes (each in GenBank flatfile format)
- DNA sequence alignments of individual genome regions (FASTA format)
- Concatenation of all individual DNA sequence alignments (FASTA and NEXUS format)
# Alignment software
apt install mafft
# Python dependencies
apt install python3-biopython
apt install python3-coloredlogs
apt install python3-requests
# Installation
pip install git+https://github.com/michaelgruenstaeudl/PlastomeBurstAndAlign.gitIf current working directory within plastburstalign, execute the package via:
python -m plastburstalignFrom within Python, execute the package functions via:
from plastburstalign import PlastomeRegionBurstAndAlign
burst = PlastomeRegionBurstAndAlign()
burst.execute()Individual components can be used as well. For example, to use the class MAFFT by itself (e.g., instantiate a configuration of MAFFT that will execute with 1 thread; institute another that will execute with 10 threads), type:
from plastburstalign import MAFFT
mafft_1 = MAFFT()
mafft_10 = MAFFT({"num_threads": 10})The plastid genome is a mosaic of individual genome regions, with many of its genes consisting of multiple exons. To align genes based on their amino acid sequence information, all exons of a gene must be extracted and concatenated prior to alignment. plastburstalign conducts this exon splicing through an automated process that differentiates between cis- or trans-spliced genes: the exons of cis-spliced genes are adjacent to each other, those of trans-spliced genes are not. The software concatenates the exons of any cis-spliced gene in place (i.e., no repositioning of the exons necessary). The exons of any trans-spliced gene (e.g., rps12), by contrast, undergo a two-step repositioning procedure before being concatenated. First, groups of contiguous exons are formed based on their location information: if an exon is adjacent to or even overlaps with another exon of the same gene name, they are merged. Second, exons of the same gene name are merged at the location of the first exon occurrence.
Due to the size and complexity of large DNA sequence alignments, individual genome regions can barely be removed from a concatenated sequence alignment; instead, any user-specified exclusion of a genome region must be performed before the actual sequence alignment. plastburstalign contains two functions for such an exclusion: commandline-parameter exclude_region excludes any user-specified region by exact name match from the dataset; commandline-parameter exclude_fullcds removes entire user-specified genes as well as any introns inside, and any intergenic spacers immediately adjacent to, the specified genes from the dataset.
The names of all tRNAs are automatically standardized across the input genomes to counteract the accumulation of idiosyncratic gene names. tRNAs are often labeled differently by different researchers. For example, researcher A may label tRNAs with both amino acid abbreviations and anticodons (e.g., trnA-Leu-UAA), whereas researcher B may label them with the respective anticodons only (e.g., trnA-Leu). Similarly, researcher C may label tRNA genes with lower-case anticodons (e.g., trnA-uaa) but researcher D with upper-case anticodons (e.g., trnA-UAA). Differences in tRNA gene names may also originate from the idiosyncratic use of dashes versus underscores (e.g., trnA-UAA versus trnA_UAA). Leaving the names of tRNAs that code for the same gene unadjusted and, thus, incongruent across different input genomes risks the artificial increase in the number of unique genes, introns, and intergenic spacers in the dataset.
To ensure that only homologous genes are grouped together and aligned, plastburstalign automatically standardizes tRNA gene names across input genomes. Specifically, the software homogenizes incongruent tRNAs gene names to a single format: tRNAabbreviation_anticodon (e.g., trnA_UAA). This format is (i) the most commonly used tRNA naming scheme among plastid genomes and (ii) the least problematic scheme for nucleotide sequence alignment operations, which typically interpret dashes as sequence characters. During the standardization operations, plastburstalign utilizes the three-letter amino acid abbreviations and the anticodon definitions of translation table 11 of the International Nucleotide Sequence Database Collaboration (INSDC). tRNAs with more than one possible codon but for which neither amino acid nor anticodon abbreviations are given in the gene name (e.g., trnA can be any of the following: trnA_UAA, trnA_CAA, trnA_AAG, trnA_GAG, trnA_UAG, and trnA_CAG), by contrast, are not changed by plastburstalign to avoid the incorrect designations.
As a side effect, the automatic standardization of tRNA gene names also decreases the number of annotated genome regions that need to be removed from the dataset for not reaching the minimum number of taxa defined. Without the standardization, the intergenic spacer between the genes trnA_CAA and ndhB, for example, may be grouped under two different names (e.g., trnA_CAA_ndhB and trnA_caa_ndhB), with the latter group being less common and eventually removed from the dataset for not reaching the minimum number of taxa. By implementing a gene name standardization, the same intergenic spacer is grouped under only one name (i.e., trnA_CAA_ndhB) and not discarded. Preliminary tests indicated that the number of annotated genome regions that were removed due to not reaching the minimum number of taxa was decreased by approximately 25% through the tRNA gene name standardization.
cd benchmarking
# CDS
python test_script_cds.py benchmarking1 5
# INT
python test_script_int.py benchmarking1 5
# IGS
python test_script_igs.py benchmarking1 5- Dataset
benchmarking1.tar.gz: all Asteraceae (n=155) listed in Yang et al. 2022 - Dataset
benchmarking2.tar.gz: all monocots (n=733) listed in Yang et al. 2022 - Dataset
benchmarking3.tar.gz: all angiosperms (n=2585) listed in Yang et al. 2022
See this document
See this document