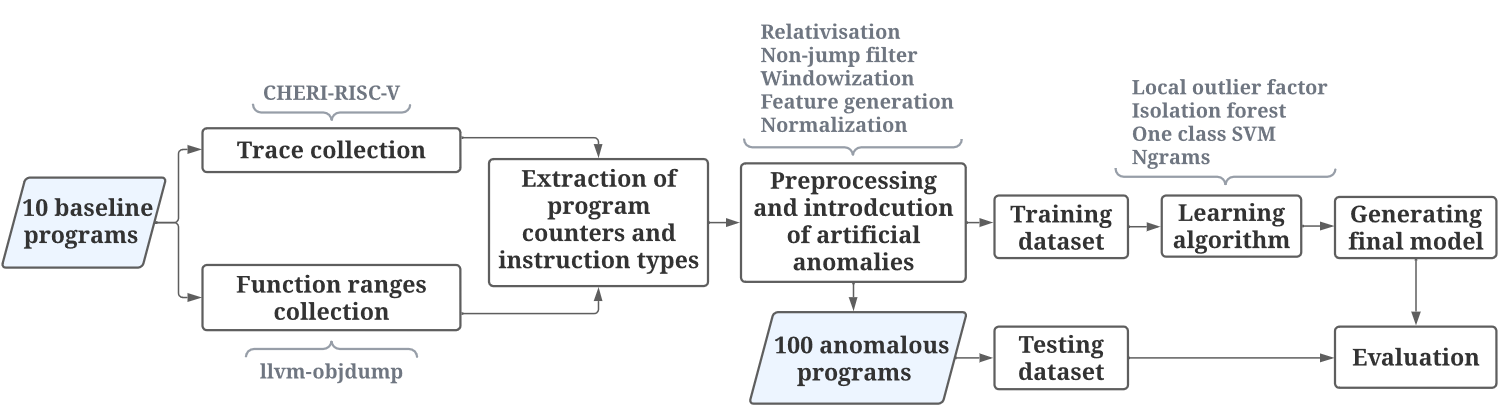
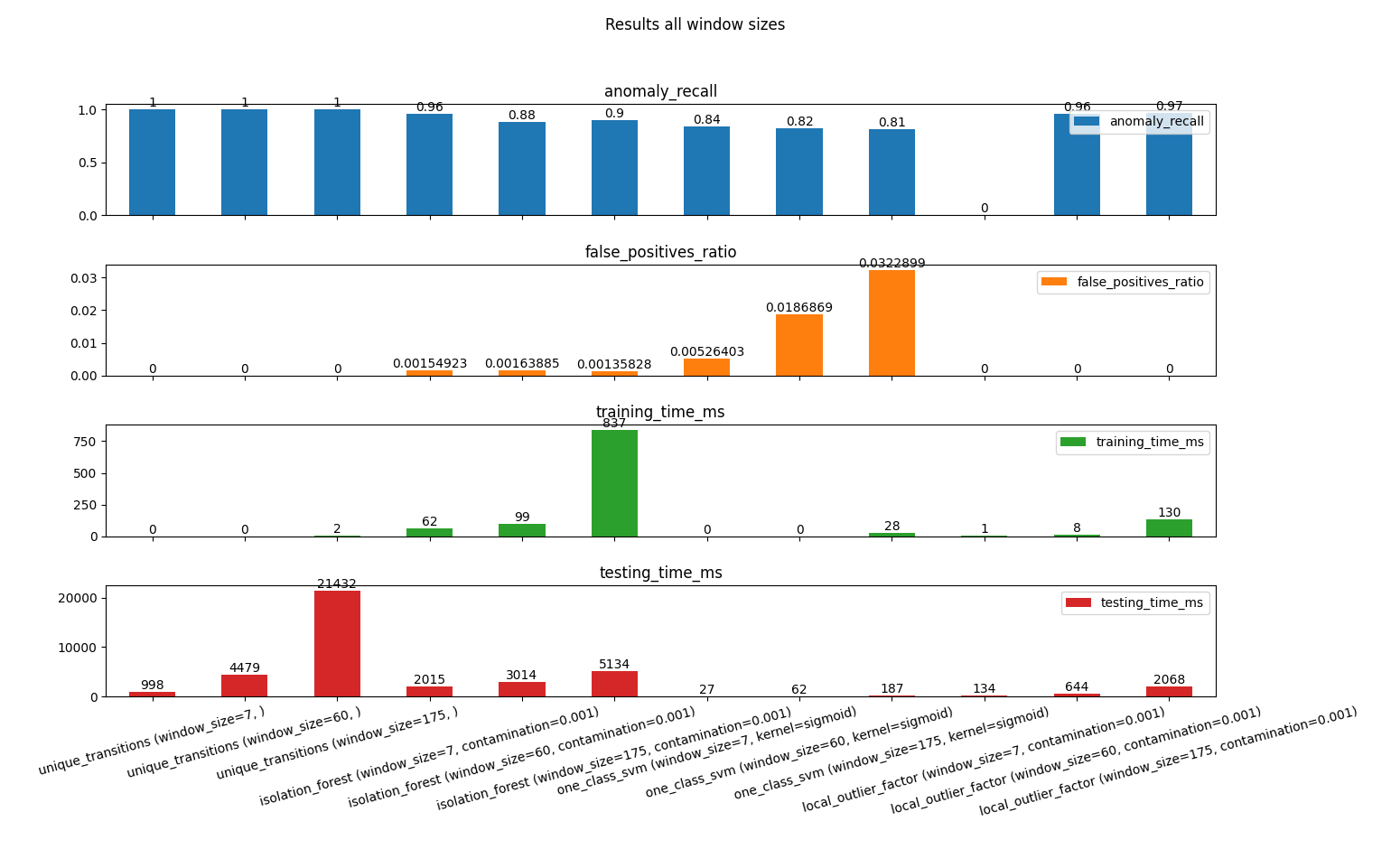
The goal of this project is to compare different methods of anomaly detection in program behaviour.
To run the comparison we can use:
python ./scripts/compare_classification_methods.py -n ./log_files/paper/csv/normal*.csv
# We can include the json file with function ranges if we wish (it does not affect detection methods, it is only supplied for plotting purposes)
python ./scripts/compare_classification_methods.py -n ./log_files/paper/csv/normal*.csv --function-ranges ./log_files/paper/*jsonInput csv files with baseline program data have two columns: program counters (in hexadecimal) and instruction types (strings). Example input file:
11FB2,addi
11FB4,sd
11FB6,sd
11FB8,addi
11FBA,mv
11FBC,sd
11FC0,sw
11FC4,auipc
11FC8,jalr
...
Abnormal files are generated artificially by copying normal input files and modifying random sections (one section per file).
Input files may be obtained in various ways (e.g. running GDB). Input files from ./log_files/paper/csv directory were obtained from Qemu emulator running CHERI-RISC-V, using qtrace -u exec ./stack-mission and then processing the collected trace log file (e.g. normal_1.log) by running the following commands:
# Obtaining output of llvm-objdump of the stack-mission program.
/tools/RISC-V/emulator/cheri/output/sdk/bin/llvm-objdump -sSD stack-mission > stack-mission-llvm-objdump.txt
# Extracting function ranges from the ".text" section of llvm-objdump output and storing it in json file.
./extract_function_ranges_from_llvm_objdump.py stack-mission-llvm-objdump.txt -o stack-mission-function-ranges.json
# Parsing the trace log using function ranges (to avoid any trace other than from the program itself, e.g. ignoring library code)
./parse_qtrace_log normal_1.log --function-ranges stack-mission-function-ranges.json -o normal_1.csvThe best way to start is to copy and rename an already implemented method. We can duplicate and rename scripts/isolation_forest/isolation_forest.py into scripts/new_method/new_method.py, and rename the class inside it into New_Method. At this point the code inside the file will look like this:
# <bunch of unused imports>
from detection_model import Detection_Model
# all detection methods must inherit from the Detection_Model class (which provides evaluation consistency)
class New_Method(Detection_Model):
def __init__(self, *args, **kwargs):
''' We can initialize our detection method however we want, in this case
it creates "model" member holding reference to IsolationForest from scikit-learn.
It isn't required to have "model" member name or any members at all being initialized. '''
self.model = IsolationForest(n_estimators=100, random_state=0, warm_start=True, *args, **kwargs)
def train(self, normal_windows, **kwargs):
# normal_windows is a 2D numpy array where each row contains input features of a single example
self.model.fit(normal_windows)
def predict(self, abnormal_windows):
''' This method must return a list of boolean values,
one for each row of "abnormal_windows" 2D numpy array. '''
# abnormal_windows is a 2D numpy array where each row contains input features of a single example
return [i==-1 for i in self.model.predict(abnormal_windows)]After implementing custom logic inside __init__, train, and predict, we have to import the new method inside compare_classification_methods.py by adding the following line where other methods are imported:
# FORMAT: from dir_name.python_file_name import class_name
from isolation_forest.isolation_forest import Isolation_Forest
from new_method.new_method import New_MethodThen inside compare_classification_methods.py we have add the new method to the anomaly_detection_models list:
anomaly_detection_models = {
# keys correspond to config file section names
# values are classes (that inherit from Detection_Model class,
# and must implement "train" and "predict",
# Detection_Model has a common evaluate_all method)
'Isolation forest' : (Isolation_Forest, {'contamination':0.001}),
'New method' : (New_Method, {'constructor_kwarg1': 5.0, 'constructor_kwarg2': 'str_value'}),
# Notice that we can add more than one configurations of the same method:
'New method' : (New_Method, {'constructor_kwarg1': 100.0, 'constructor_kwarg2': 'another_value'})
}The last step is to add new section (with name corresponding to the key of anomaly_detection_models list above) in the configuration file (compare_classification_methods_config.ini):
[Isolation forest]
train_using_abnormal_windows_too=False
normalize_dataset=True
[New method]
train_using_abnormal_windows_too=False
normalize_dataset=TrueAt this point the new detection method should be ready. When we run the comparison script, it will be trained, tested and included in plots.