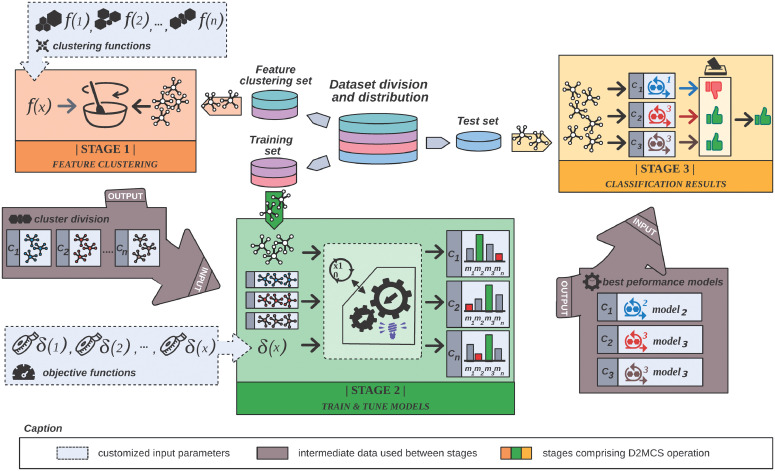
R framework to automatically predict the biological activity of chemical compounds regarding to theirs chemical substructures
- r-base
- r-base-core
Download from CRAN Project: https://cran.r-project.org/bin/windows/
| R Libraries | ||||
|---|---|---|---|---|
| R6 | car | plyr | dplyr | caret |
| mltools | here | parallel | RWeka | devtools |
| ModelMetrics | tools | graphics | gridExtra | grid |
| lattice | rJava | FSelector | BBmisc | ggalt |
| tictoc | ggrepel | ggplot2 | reshape2 |
source("sources.R")
data <- Dataset$new( filepath ="<path_to_dataset_file>", header=TRUE,
sep="\t",skip = 1, normalize.names=TRUE, classIndex = 1 )
data$executePartition(4)
trFunction <- TwoClass$new( method = "cv", number = 10, savePredictions = "final",
classProbs = TRUE, allowParallel = TRUE, verboseIter = FALSE)
fisherC <- BinaryFisherClustering$new(dataset = data$getSubset(1,2), maxClusters = 50)
fisherC$execute(positive.class = "<value_positive_class>")
train.subset <- fisherC$createSubset( subset = data$getSubset(c(2,3)) )
test.subset <- data$getSubset(4)
Benchmarking <- D2MCS$new( path = "models/BinaryFisherCluster", trainFunction = trFunction )
Benchmarking$train( train.set = train.subset, metric = "MCC" )
classify <- Benchmarking$classify( test.set = test.subset), voting.scheme = ClassWeightedVoting$new(),
positive.class = "<value_positive_class>" )
classify$computePerformance( ob = test.subset$getClass(), list(MCC$new(), PPV$new(), Accuracy$new()) )
Output visualization example
| Measure | Value | |
|---|---|---|
| 1 | MCC | 0.7858148 |
| 2 | PPV | 0.8864542 |
| 3 | Accuracy | 0.8928571 |
optimization <- Benchmarking$optimize( opt.set = test.subset,
voting.scheme = ClassWeightedVoting$new(),
opt.algorithm = list(NSGAII$new( min.function = FPFN$new(),
n.generations = 25000,
n.iteractions = 1 ),
SMSEMOA$new( min.function = FPFN$new(),
n.generations = 25000,
n.iteractions = 1 ) ),
positive.class = "<value_positive_class>" )To cite D2-MCS please use:
Ruano-Ordás, D; Yevseyeva, I, Basto-Fernandes,V; Méndez, José R; Emmerichc, Michael T.M. (2018). Improving the drug discovery process by using multiple classifier systems Expert Systems with Applications. Volume 121, pp. 292-303. Elsevier. https://doi.org/10.1016/j.eswa.2018.12.032