This repository contains the code for the constraint layer proposed in our paper titled "How Realistic Is Your Synthetic Data? Constraining Deep Generative Models for Tabular Data", which was accepted for publication at ICLR 2024.
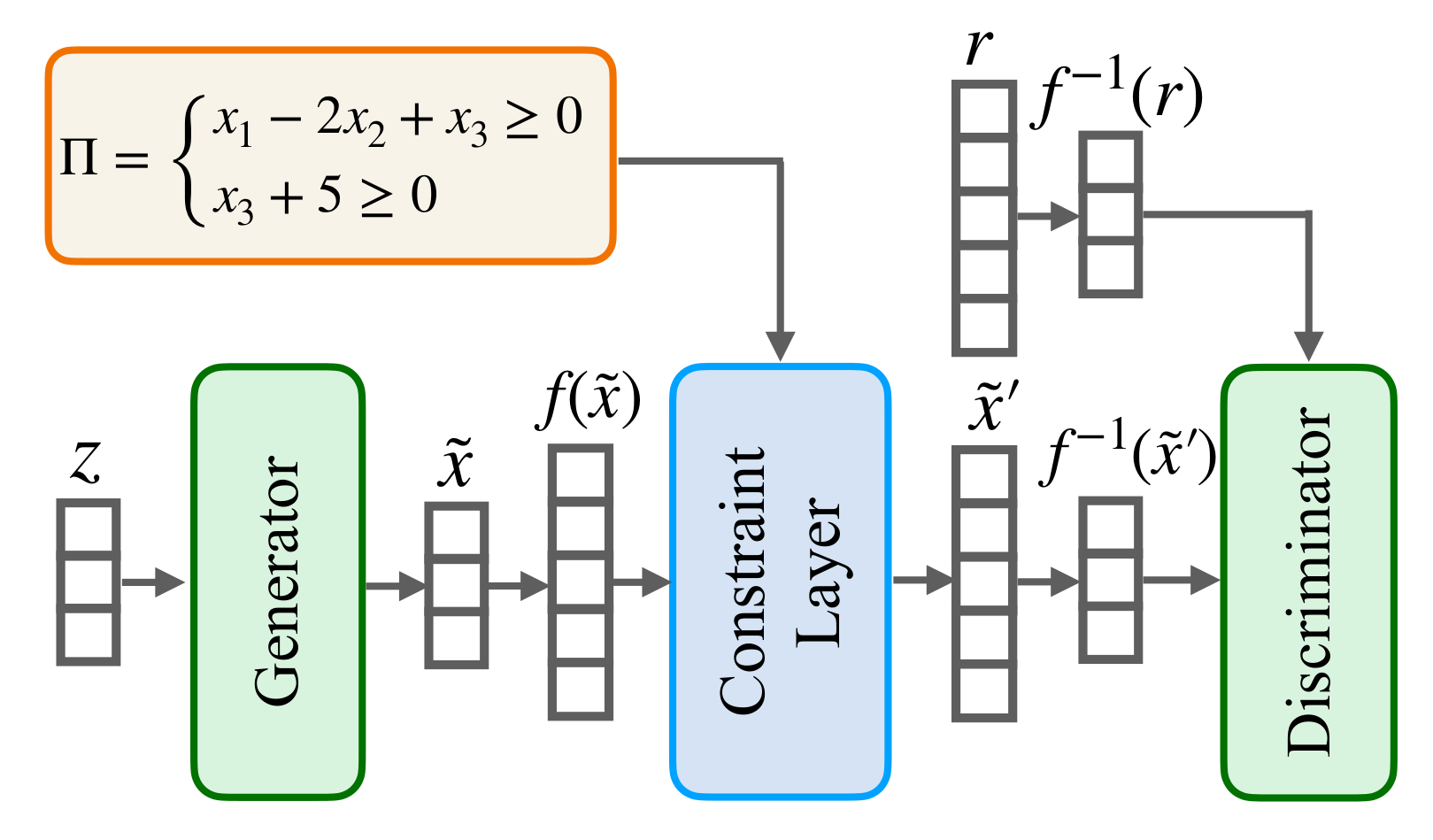
Deep Generative Models (DGMs) have been shown to be powerful tools for generating tabular data. However, they fail to comply with constraints that encode essential background knowledge on the problem at hand, which is needed to generate realistic synthetic data. In our paper, we address this limitation and show how DGMs for tabular data can be transformed into Constrained Deep Generative Models (C-DGMs), whose generated samples are guaranteed to be compliant with the given constraints. This is achieved by automatically parsing the constraints and transforming them into a Constraint Layer (CL) seamlessly integrated with the DGM, as shown below.
If you use this paper, please cite the following:
@inproceedings{cdgm2024iclr,
title = "How Realistic Is Your Synthetic Data? Constraining Deep Generative Models for Tabular Data",
author = "Mihaela Catalina Stoian and Salijona Dyrmishi and Maxime Cordy and Thomas Lukasiewicz and Eleonora Giunchiglia",
year = "2024",
booktitle = "Proceedings of the 12th International Conference on Learning Representations, ICLR 2024, Vienna, Austria, 7--11 May 2024",
month = "May",
}
As of now, the models included in this repository (on top of which we applied our constraint layer) are: CTGAN [1], TVAE [1], and TableGAN [2].
conda create -n "cdgm" python=3.10 ipython
conda activate cdgm
conda install pytorch torchvision torchaudio cpuonly -c pytorch
pip install sdv
pip install autograd
pip install wandb
pip install synthcity
To download the datasets as they were used in this paper please use this link https://figshare.com/account/articles/26830996?file=48787246. Place train, val and test splits in the approriate dataset folder under data directory.
Please refer to the appendix of our paper where we detail the datasets we used, the original source and pre-processing if any.
If needed, the scripts that we used to split the data into train, validation and testing partitions are available in the other_helper_scripts directory of this repository.
use_case="url"
eps=300
default_optimiser="adam"
default_lr=0.0010
default_bs=128
default_random_dim=100
seed=0
# unconstrained
python main_tableGAN.py ${use_case} --seed=$seed --epochs=$eps --optimiser=${default_optimiser} --lr=${default_lr} --batch_size=${default_bs} --random_dim=${default_random_dim}
# constrained
python main_tableGAN.py ${use_case} --seed=$seed --epochs=$eps --optimiser=${default_optimiser} --lr=${default_lr} --batch_size=${default_bs} --random_dim=${default_random_dim} --version="constrained" --label_ordering="corr"
use_case="url"
eps=150
default_optimiser="adam"
default_lr=0.0002
default_bs=500
default_decay=0.000001
default_pac=1
seed=0
# unconstrained
python main_ctgan.py ${use_case} --wandb_project=$wandbp --seed=$seed --epochs=$eps --optimiser=${default_optimiser} --generator_lr=${default_lr} --discriminator_lr=${default_lr} --batch_size=${default_bs} --generator_decay=${default_decay} --discriminator_decay=${default_decay} --pac=${default_pac}
# constrained
python main_ctgan.py ${use_case} --wandb_project=$wandbp --seed=$seed --epochs=$eps --optimiser=${default_optimiser} --generator_lr=${default_lr} --discriminator_lr=${default_lr} --batch_size=${default_bs} --generator_decay=${default_decay} --discriminator_decay=${default_decay} --pac=${default_pac} --version="constrained" --label_ordering="random"
use_case="url"
seed=0
eps=150
bs=70
l2scale=0.0002
loss_factor=2
# unconstrained
python main_tvae.py ${use_case} --wandb_project=$wandbp --seed=$seed --epochs=$eps --batch_size=${bs} --l2scale=${l2scale} --loss_factor=${loss_factor}
# constrained
python main_tvae.py ${use_case} --wandb_project=$wandbp --seed=$seed --epochs=$eps --batch_size=${bs} --l2scale=${l2scale} --loss_factor=${loss_factor} --version="constrained" --label_ordering="random"
References
[1] Lei Xu, Maria Skoularidou, Alfredo Cuesta-Infante, and Kalyan Veeramachaneni. Modeling tabular data using conditional GAN. In Proceedings of Neural Information Processing Systems, 2019.
[2] Noseong Park, Mahmoud Mohammadi, Kshitij Gorde, Sushil Jajodia, Hongkyu Park, and Young-min Kim. Data synthesis based on generative adversarial networks. Proceedings of the VLDB Endowment, 11, 2018.