The goal of climateR-catalogs is to “automate” a collection of data
catalogs usable with R(e.g. terra, climateR, gdalio, geoknife,
stars) and in Python (e.g. gdptools)
The catalog(s) are built using targets in this repo, and deployed as
JSON/parquet/rds artifacts at
“https://mikejohnson51.github.io/climateR-catalogs/catalog.{ext}”.
The catalog will hope to support 4 types of data source and will limit itself to non-authenticated public data assets. The included types will be:
- Aggregated NetCDF (via OpenDap)
- Non aggregated NetCDF (via OpenDap)
- tif/COG/vrt hosted by s3/FTP/ect
- STAC catalog (WIP)
Targets will be used in an attempt to ensure maintenance is possible. The interactive version of the latest run can be seen here here
To align the 4 categories of data a WIP schema can be found here
The catalog looks like this on 2023-03-08 reading local version (same as
pushed one found in docs)
cat = read_parquet("docs/catalog.parquet")
# Unique datasets
nrow(cat)
#> [1] 107857
# Unique products
length(unique(cat$asset))
#> [1] 4653Here is a minimal example with the base information added:
pacman::p_load(dplyr, climateR, AOI, terra)
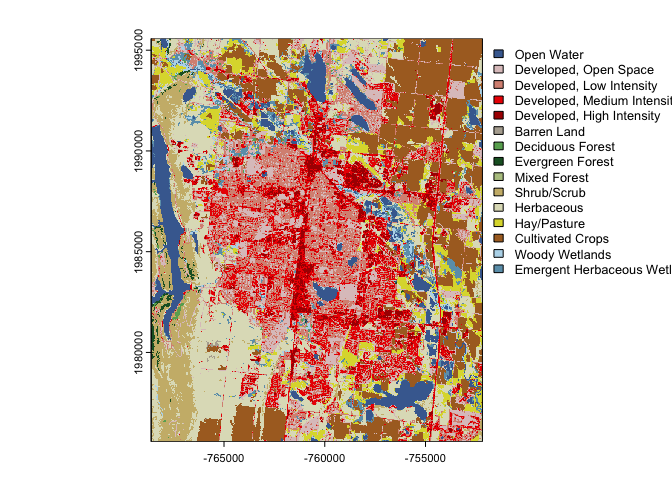
nlcd = filter(cat, id == "NLCD", asset == '2019 Land Cover L48')
t(nlcd)
#> [,1]
#> id "NLCD"
#> asset "2019 Land Cover L48"
#> URL "/vsicurl/https://storage.googleapis.com/feddata-r/nlcd/2019_Land_Cover_L48.tif"
#> type "vrt"
#> varname "Land_Cover"
#> variable "Land_Cover"
#> description "USGS NLCD Land Cover 2019 L48"
#> units ""
#> model NA
#> ensemble NA
#> scenario NA
#> T_name NA
#> duration NA
#> interval NA
#> nT NA
#> X_name NA
#> Y_name NA
#> X1 "-2493045"
#> Xn "2342655"
#> Y1 "-2493045"
#> Yn "3310005"
#> resX "30"
#> resY "30"
#> ncols "161190"
#> nrows "104424"
#> crs "+proj=aea +lat_0=23 +lon_0=-96 +lat_1=29.5 +lat_2=45.5 +x_0=0 +y_0=0 +datum=WGS84 +units=m +no_defs"
#> toptobottom NA
#> tiled ""(output = dap(catalog = nlcd, AOI = aoi_get("Fort Collins")))
#> $`2019 Land Cover L48`
#> class : SpatRaster
#> dimensions : 667, 548, 1 (nrow, ncol, nlyr)
#> resolution : 30, 30 (x, y)
#> extent : -768615, -752175, 1975575, 1995585 (xmin, xmax, ymin, ymax)
#> coord. ref. : Albers Conical Equal Area
#> source(s) : memory
#> color table : 1
#> categories : NLCD Land Cover Class, Histogram, Red, Green, Blue, Opacity
#> name : NLCD Land Cover Class
#> min value : Open Water
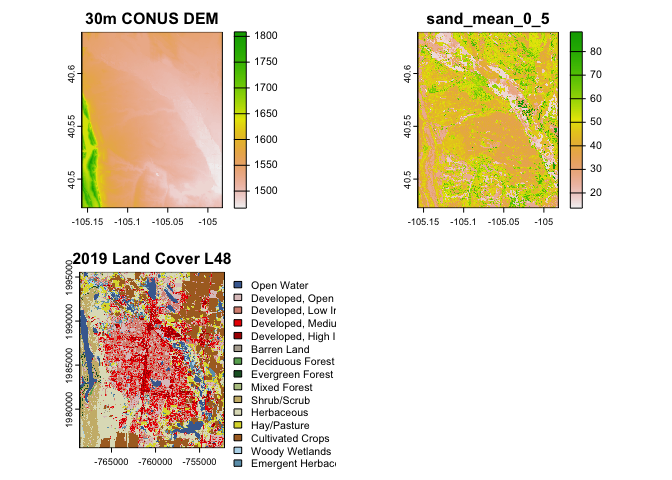
#> max value : Emergent Herbaceous WetlandsThis is still a little clunky but you can pass multi-row “catalog” data.frames straight to dap! For example say you want soil sand content, land cover and elevation for the city of Fort Collins:
multilayer = filter(cat, asset %in% c("2019 Land Cover L48", "30m CONUS DEM", "sand_mean_0_5"))
output = sapply(1:nrow(multilayer), function(x){
dap(catalog = multilayer[x,], AOI = aoi_get("Fort Collins"))
})dap_resource = filter(cat,
id == 'bcca',
variable == 'tasmin',
model == 'MPI-ESM-LR',
ensemble == 'r1i1p1',
scenario == "historical")
t(dap_resource)
#> [,1]
#> id "bcca"
#> asset "historical"
#> URL "https://cida.usgs.gov/thredds/dodsC/cmip5_bcca/historical"
#> type "opendap"
#> varname "BCCA_0-125deg_tasmin_day_MPI-ESM-LR_historical_r1i1p1"
#> variable "tasmin"
#> description "Daily Minimum Near-Surface Air Temperature"
#> units "C"
#> model "MPI-ESM-LR"
#> ensemble "r1i1p1"
#> scenario "historical"
#> T_name "time"
#> duration "1950-01-01 12:00:00/2005-12-31 12:00:00"
#> interval "1 days"
#> nT "20454"
#> X_name "longitude"
#> Y_name "latitude"
#> X1 "-124.6875"
#> Xn "-67.0625"
#> Y1 "25.1875"
#> Yn "52.8125"
#> resX "0.125"
#> resY "0.125"
#> ncols "462"
#> nrows "222"
#> crs "+proj=longlat +a=6378137 +f=0.00335281066474748 +pm=0 +no_defs"
#> toptobottom "TRUE"
#> tiled "T"
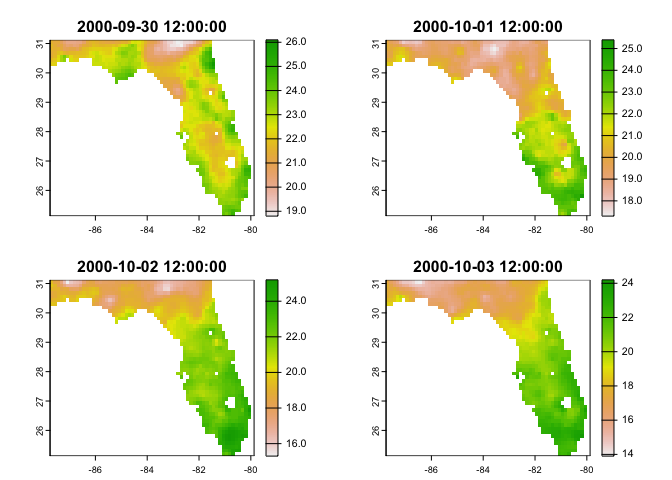
data = dap(URL = dap_resource$URL,
AOI = aoi_get(state = "FL"),
varname = dap_resource$varname,
startDate = "2000-10-01",
endDate = "2000-10-04")
#> source: https://cida.usgs.gov/thredds/dodsC/cmip5_bcca/historical
#> varname(s):
#> > BCCA_0-125deg_tasmin_day_MPI-ESM-LR_historical_r1i1p1 [C] (Daily Minimum Near-Surface Air Temperature)
#> ==================================================
#> diminsions: 63, 48, 4 (names: longitude,latitude,time)
#> resolution: 0.125, 0.125, 1 days
#> extent: -87.75, -79.88, 25.12, 31.12 (xmin, xmax, ymin, ymax)
#> crs: +proj=longlat +a=6378137 +f=0.00335281066474748 +p...
#> time: 2000-09-30 12:00:00 to 2000-10-03 12:00:00
#> ==================================================
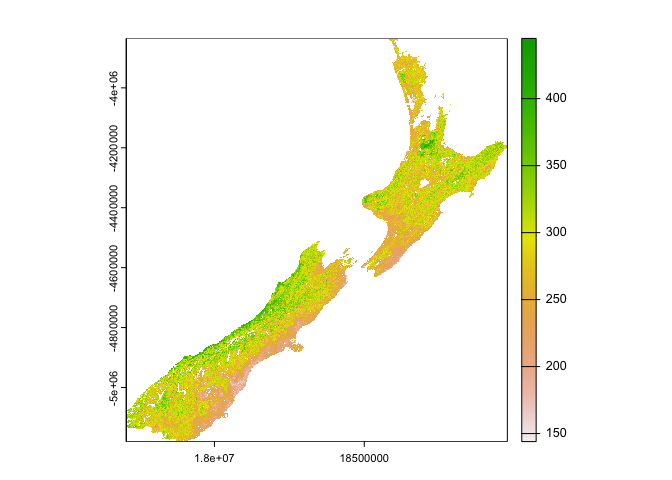
#> values: 12,096 (vars*X*Y*T)nz_soil = filter(cat, id == "ISRIC Soil Grids", variable == 'silt_0-5cm_mean')
data = dap(URL = nz_soil$URL, AOI = aoi_get(country = "New Zealand"))