This repository provides a PyTorch implementation of our work accepted by ISBI 2024 --> [arXiv]
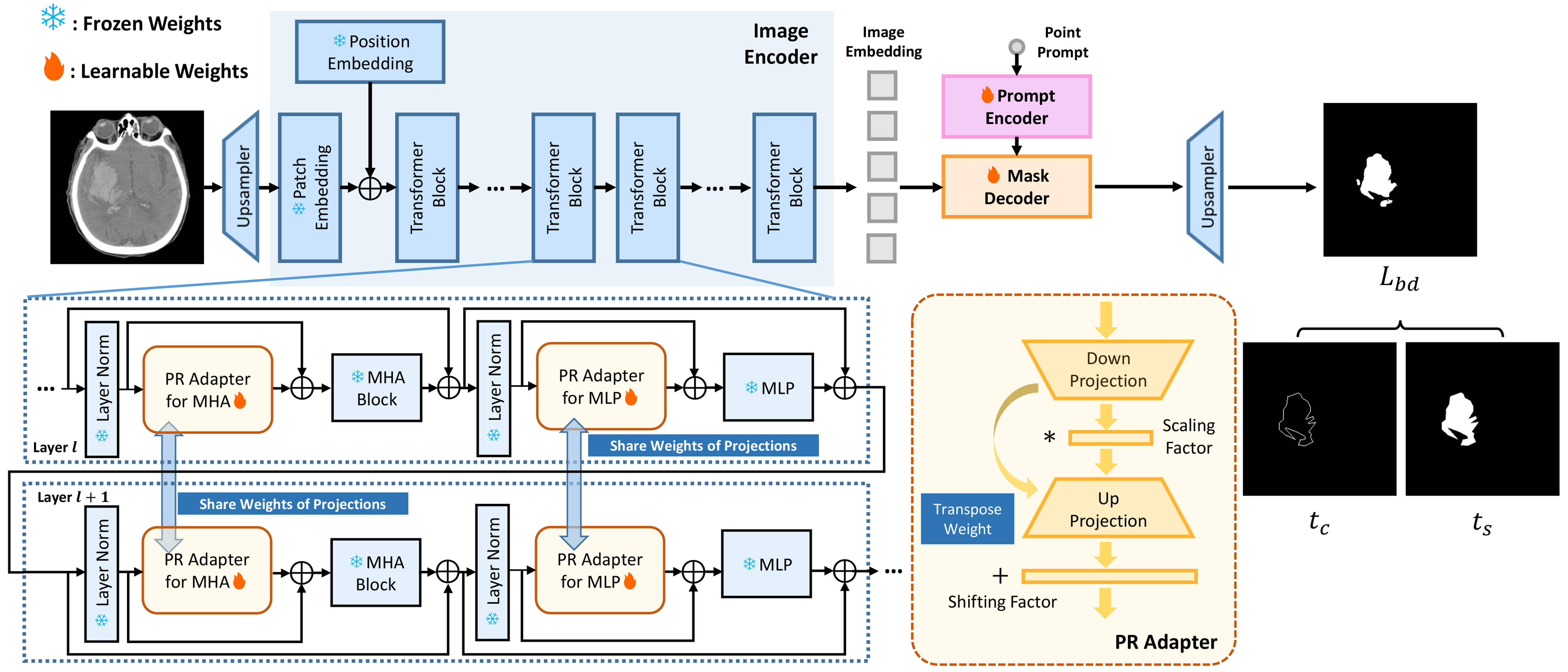
- Model: SAMIHS: A parameter-efficient fine-tuning (PEFT) method
- Task: To adapt the Segment Anything Model (SAM) to intracranial hemorrhage segmentation.
- Ideas: The parameter-refactoring adapters and boundary-sensitive loss are incorporated in SAMIHS to improve both efficiency and accuracy.
- 2023.11.13: Code released.
$ git clone https://github.com/mileswyn/SAMIHS.git
$ cd SAMIHS/
$ pip install requirements.txtWe use checkpoint of SAM in vit_b version. Please download the pre-trained model and place it at pretrained/sam_vit_b_01ec64.pth.
- We have evaluated our method on two publicly-available datasets: BCIHM Instance.
- After downloading the datasets, you can follow the
utils/preprocess.pyto save the slice in.npyformat, and read them with the information in pathdataset/excel/. - The relevant information of your data should be set in ./utils/config.py .
If you have already arranged your data, you can start training your model.
cd "/home/... .../SAMIHS/"
python train.py -task <your dataset name> -sam_ckpt <pre-trained model path> -fold <fold number>
After finishing training, you can start testing your model.
python test.py -task <your dataset name> -sam_ckpt <pre-trained model path> -fold <fold number>
Before testing, don't forget modify the "load_path" (the path of your trained model) in [./utils/config.py].
If our SAMIHS is helpful to you, please consider citing our paper:
@article{wang2023samihs,
title={SAMIHS: Adaptation of Segment Anything Model for Intracranial Hemorrhage Segmentation},
author={Wang, Yinuo and Chen, Kai and Yuan, Weimin and Meng, Cai and Bai, XiangZhi},
journal={arXiv preprint arXiv:2311.08190},
year={2023}
}
- A lot of code is modified from SAMUS.