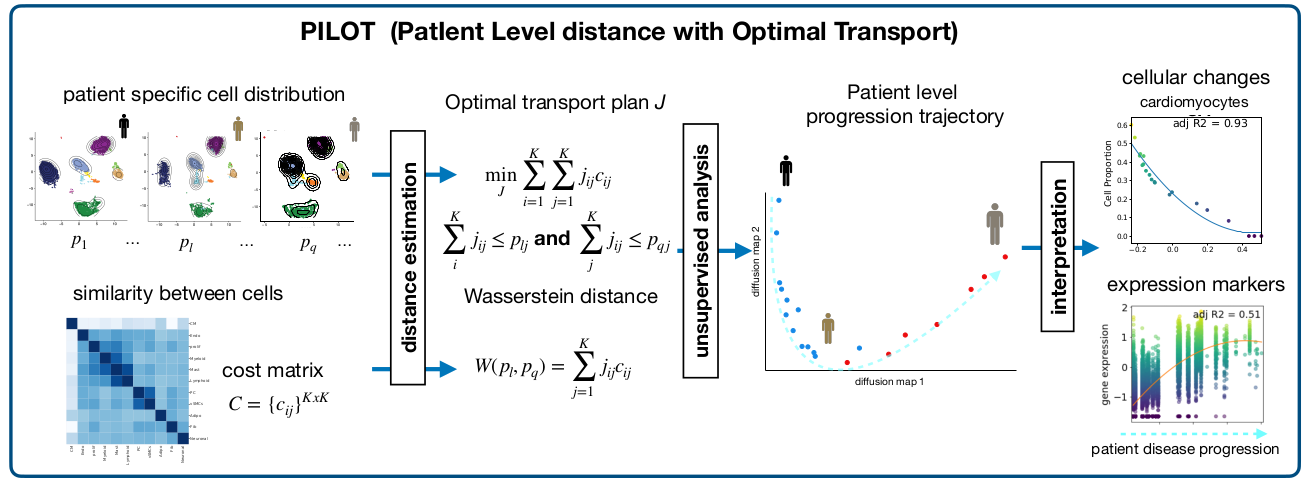
PILOT uses optimal transport to compute the Wasserstein distance between two single single-cell experiments. This allows us to perform unsupervised analysis at the sample level and to uncover trajectories associated with disease progression. Moreover, PILOT provides a statistical approach to delineate non-linear changes in cell populations, gene expression and tissues structures related to the disease trajectories. We evaluate PILOT and competing approaches in disease single-cell genomics and pathomics studies with up to 1.000 patients/donors and millions of cells or structures. Results demonstrate that PILOT detects disease-associated samples, cells, and genes from large and complex single-cell and pathomics data.
Current version for PILOT is 0.1.0
The easiest way to install PILOT and the required packages is using pip,
git clone https://github.com/CostaLab/PILOT
cd PILOT
pip install .
For applying PILOT to your data, we provide tutorials here, you just need to after installation, follow the same path in the Tutorial. Please just make sure that you need to extract needed materials as inputs for PILOT. You can see the required data and its structure in Datasets.