 by Fabian Laumer, Gabriel Fringeli, Alina Dubatovka, Laura Manduchi, Joachim M. Buhmann
by Fabian Laumer, Gabriel Fringeli, Alina Dubatovka, Laura Manduchi, Joachim M. Buhmann
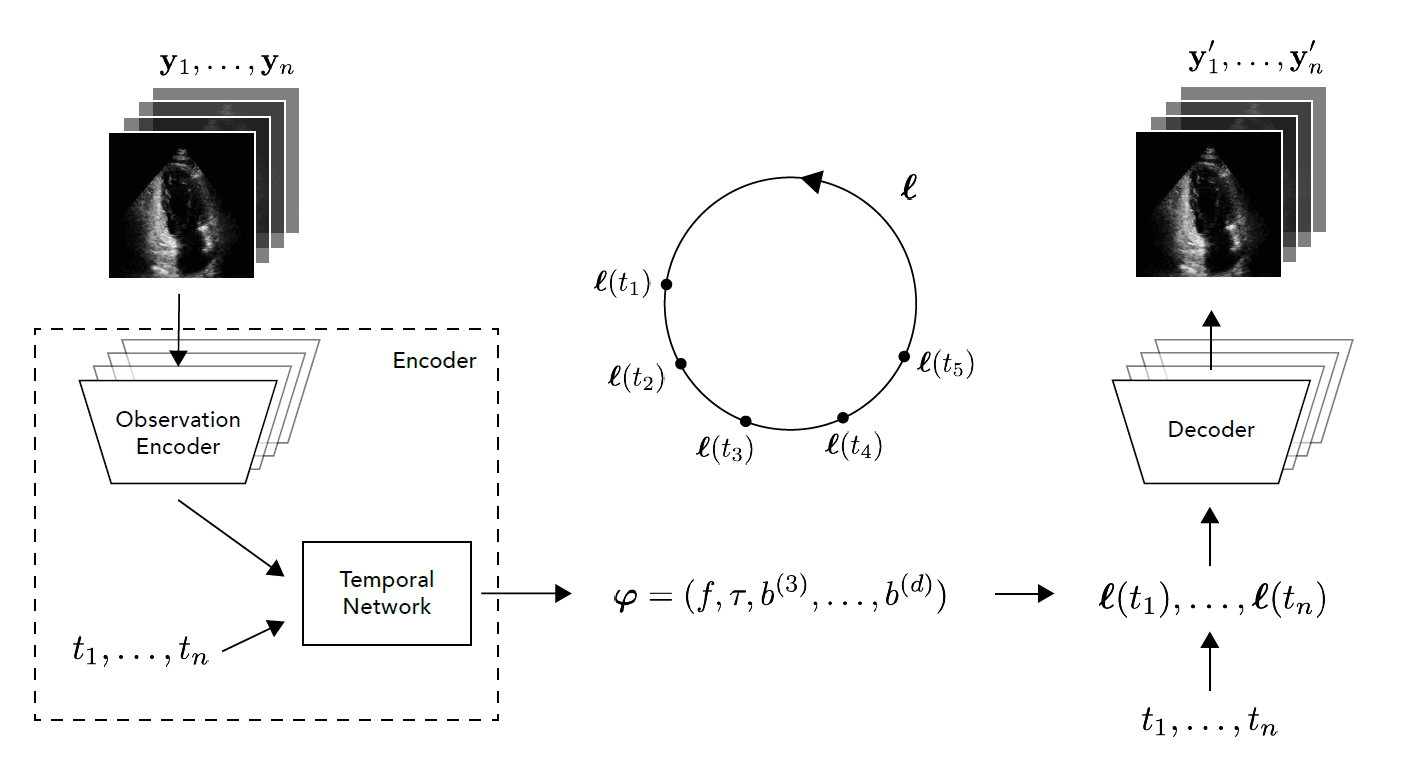
This repository contains the accompanying code to our paper DeepHeartBeat: Latent trajectory learning of cardiac cycles using cardiac ultrasounds, which presents a novel autoencoder-based framework for learning human interpretable representations of cardiac cycles from cardiac ultrasound data.
We provide the pre-trained TensorFlow models used in the experiments of our paper, which includes:
- Echocardiogram models trained on the EchoNet-Dynamic dataset
- Single Lead ECG model trained on the PhysioNet Computing in Cardiology Challenge 2017 dataset
The models can be loaded in the following way:
from utils import *
# EchoNet-Dynmaic
model = load_echonet_dynamic_model(i) # with i in [0, 1, 2, 3, 4]
# PhysioNet
model = load_physionet_model()
If you are interested in the model architectures and training procedures used for fitting either data type, you may consider the model definitions contained in models/.
The folder experiments/ contains all the code required to reproduce the experiments of the paper. In order to be able to run the experiments you need to place the EchoNet-Dynamic data in the path data/EchoNet-Dynamic/ and the PhysioNet data in the path data/physionet.org/.
The project contains the following experiments:
experiments/train_echo.py: Fits five DeepHeartBeat models to the EchoNet-Dynamic echogram dataset and saves the model weights intrained_models/.experiments/eval_echo.py: Evaluation of the DeepHeartBeat models trained inexperiments/train_echo.py. This includes heart rate detection, semantic alignment, and the prediction of the ejection fraction of the left ventricle.experiments/rnmf_heart_rates.py: Heart rate detection for the EchoNet-Dynamic echocardiograms using Rank-2 Robust Non-negative Matrix Factorisation (RNMF).
experiments/train_ecg.py: Fits a single DeepHeartBeat model to the PhysioNet single lead ECG dataset and saves the model weights intrained_models/.experiments/eval_ecg.py: Evaluation of the DeepHeartBeat model trained inexperiments/train_ecg.py. This includes anomaly detection (noise detection) and atrial fibrillation (AF) detection.
To run any of the experiments you can use the following command:
$ python run.py [EXPERIMENT NAME]
For example to trun the Echo evaluation you may invoke
$ python run.py eval_echo
When using this code, please cite:
@InProceedings{pmlr-v136-laumer20a,
title = "DeepHeartBeat: Latent trajectory learning of cardiac cycles using cardiac ultrasounds",
author = "Laumer, Fabian and Fringeli, Gabriel and Dubatovka, Alina and Manduchi, Laura and Buhmann, Joachim M.",
booktitle = "Proceedings of the Machine Learning for Health NeurIPS Workshop",
pages = "194--212",
year = "2020",
editor = "Emily Alsentzer and Matthew B. A. McDermott and Fabian Falck and Suproteem K. Sarkar and Subhrajit Roy and Stephanie L. Hyland",
volume = "136",
series = "Proceedings of Machine Learning Research",
month = "11 Dec",
publisher = "PMLR",
pdf = "http://proceedings.mlr.press/v136/laumer20a/laumer20a.pdf",
url = "http://proceedings.mlr.press/v136/laumer20a.html"
}