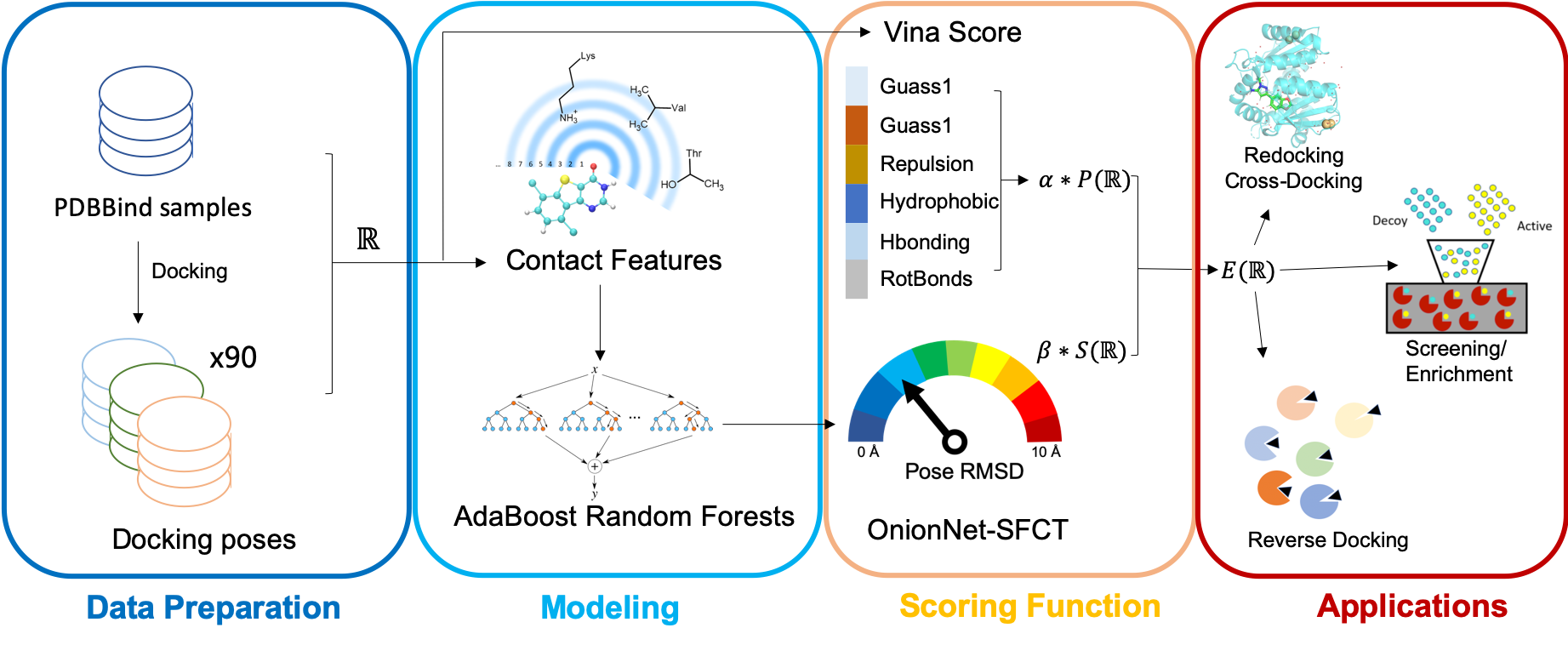
Key points of OnionNet-SFCT:
- A machine learning model (OnionNet-SFCT) is come up to correct the scoring by physical or empirical scoring function (Vina score).
- The model shows good performance on docking related tasks (redocking and cross-docking).
- The screening accuracies are almost doubled when Vina score is equipped with OnionNet-SFCT.
- The combination of Vina score and OnionNet-SFCT could be applied for reverse screening.
- OnionNet-SFCT captures certain short-range polar interactions between the protein and the ligand.
OnionNet-SFCT is only a scoring term, should be used in combination with Vina/Qvina/iDock, and the docking accurcies may vary, and should be compared to the original scoring term (Vina score, or Qvina score). If the docking engine (Vina/Qvina/iDock) fails to generate the right poses, it is not possible for OnionNet to select the near-native pose.
There will be a webserver available soon on the Zcloud platform for testing.
Now there is a docker image available for those wishing an easier solution:
https://hub.docker.com/repository/docker/hotwa/onionnet_sfct
Many thanks to @hotwa :-) !!!
Rescoring the docking results generated by AutoDock Vina.
cd example/
python scorer.py -r receptor.pdb -l active_1_result.pdbqt \
-o scores.dat --model ../data/sfct.model \
--ref crystal_active.mol2
Arguments explained:
-r : the input receptor (protein) file, pdb or pdbqt format
-l : the vina output pdbqt file containing vina scores and docking poses
-o : the output scores
--model : the scoring model file (AdaBoostRegressor pickle file)
-w : the weight factor w, default is 0.5
--ref : the reference crystal ligand file, mol2 format or sdf format
#pose_index origin_score combined_score sfct RMSD(Angstrom)
0 -8.200 -2.067 4.066 inf
1 -8.000 -1.389 5.221 inf
2 -7.900 -1.385 5.130 inf
3 -7.900 -1.469 4.963 inf
4 -7.800 -1.309 5.182 inf
5 -7.700 -1.224 5.251 inf
6 -7.700 -1.199 5.302 inf
7 -7.700 -1.478 4.745 inf
8 -7.600 -1.141 5.317 inf
9 -7.500 -1.264 4.973 inf
10 -7.500 -1.231 5.039 inf
11 -7.500 -1.053 5.393 inf
12 -7.400 -0.905 5.590 inf
13 -7.400 -1.026 5.348 inf
14 -7.400 -1.465 4.469 inf
15 -7.300 -1.041 5.218 inf
16 -7.200 -0.855 5.491 inf
17 -7.200 -0.960 5.280 inf
18 -7.100 -0.800 5.501 inf
19 -7.100 -0.945 5.209 inf
- pose_index: the pose identifier, the pose index number
- origin_score: the AutoDock Vina score in the input pdbqt file (a Vina docking result file)
- combined_score: the OnionNet-SFCT+Vina score (default weight w=0.5)
- sfct: the OnionNet-SFCT predicted score, the predicted RMSD value
- (optional) RMSD: if reference molecule (often the crystal molecule) is provided, the RMSD values of the docking poses with respect to the reference molecule are calculated. Please note that the model is designed for ligand screening, the RMSD predicted by this model is generate over-estimated, meaning that the predicted RMSD is often larger than the actual RMSD, but the trend of the RMSD of different poses should be reliable.
Liangzhen Zheng, Jintao Meng, Kai Jiang, Haidong Lan, Zechen Wang, Mingzhi Lin, Weifeng Li, Hongwei Guo, Yanjie Wei, Yuguang Mu, Improving protein–ligand docking and screening accuracies by incorporating a scoring function correction term, Briefings in Bioinformatics, 2022;, bbac051, https://doi.org/10.1093/bib/bbac051
The standard model file could found here: https://drive.google.com/file/d/1iiJvW4GBfg4D7LCuTRLKv9qnRYu5L2o5/view?usp=drive_link .
Zheng Liangzhen, astrozheng_AT_gmail.com