Dermoscopic Image Classification
This repository is corresponding to the paper Tackling Imbalance dermoscopic image classification using data augmentation and GAN and it contains the source code and implementation of published study.
Abstract
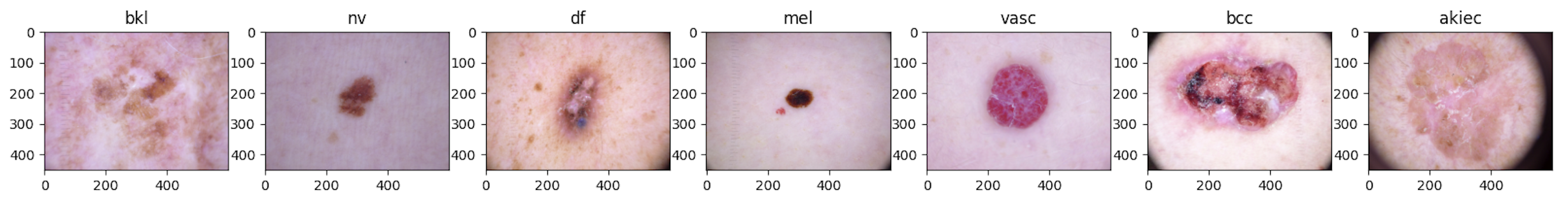
Dermoscopy is a noninvasive way to examine and diagnose skin lesions, e.g. nevus and melanoma, and is a critical step for skin cancer detection. Accurate classification of dermoscopic images can detect skin cancer at an early stage and bring social and economic impact to patients and communities. Using deep learning methods to classify dermoscopic images has shown superior performance, but existing research often overlooks the class imbalance in the data. In addition, although a handful of public datasets are available for skin cancer research, these datasets are generally not large enough for deep learning algorithms to produce accurate results. In this paper, we propose to use data augmentation and generative adversarial networks (GAN) to tackle class-imbalanced dermoscopic image classification. Our main objectives are to determine (1) how state-of-the-art fine-tuned deep learning models perform on class-imbalanced dermoscopic images, (2) whether data augmentation and GAN can help alleviate class imbalances to improve classification accuracy, and (3) which method is more effective in addressing the class imbalance. By using public datasets and a carefully designed framework to generate augmented images and synthetic images, our research provides clear answers to these questions
Quick Links
Dataset EDA
-
Data ReadMe file Data ReadMe describes the dataset used in this study and provides link to download the dataset needed to replicate our study.
-
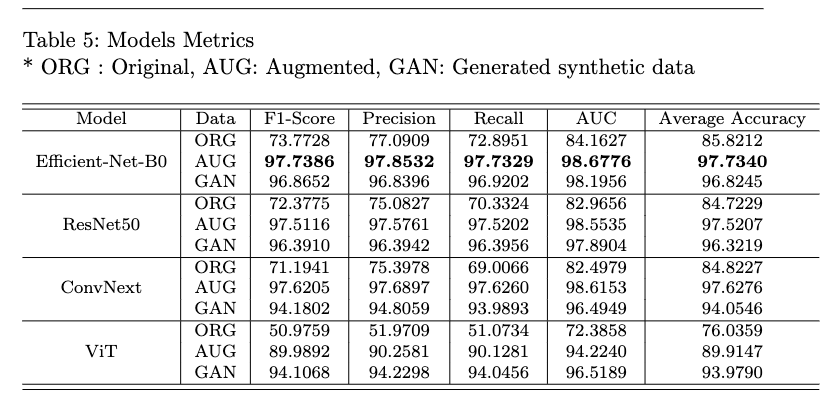
We also show data distribution after augmentation and after synthetic image generation using AC-GAN.
Models
-
Models ReadMe file contains all the information about the models used in this study.
-
Implementation and training of AC-GAN
Setup
-
Clone GitHub Repository
git clone [repo link] -
Create Environment
We used conda environment for our virtual environment
conda create -n dermcv python=3.7 -yconda activate dermcv -
Install Dependencies
pip install -r requirements.txt -
Install Pytorch with GPU Verify that you pytorch is installed and cuda is configured.
pip install torch==1.12.0+cu116 torchvision==0.13.0+cu116 torchaudio==0.12.0 --extra-index-url https://download.pytorch.org/whl/cu116
- Verify Pytorch has Cuda
import torch
torch.cuda.is_available()
Repository Structure
├── README.md
├── csv
├── data
├── logs
├── models
├── notebooks
│ ├── GAN-Augmentation.ipynb
│ └── data_explration.ipynb
├── requirements.txt
├── save
└── src
├── GAN-PyTorch.py
├── config.py
├── dataset.py
├── imports.py
├── models.py
├── train.py
└── utils.py
Running
Training
train.py contain all the training scripts while the parameters used are located in the config.py. To make changes to the parameters just change the default values in the config.py or pass it with the training script.
python train.py
Preprocessing
- Augmentation
- Generative Adversarial Networks
GAN Training
We used AC-GAN to generate Images. It needs to be trained on the minority class image that needs to be extracted from the meta file on the HAM10k meta file. Data folder should contain a single folder named HAM10k that contains all the images, while the meta files should be placed in the csv folder as train/test.
python GAN-Pytorch.py --data your-data-folder --csv_files csv-files-folder --n_epochs epochs --batch_size 64 --n_classes minority-classes
Models to be used
- Efficientnet
- ViT
- ConvNext
- ResNet50
- CNN
Contribution
@malsaidi @mjan2021