bodynavigation
Segmentation of internal organs from Computed Tomography images of- ten uses intensity and shape properties. We introduce a navigation system based on robust segmentation of body tissues like spine, body surface and lungs. Pose esti- mation of an investigated tissue can be performed using this algorithm and it also can be used as a support information for various segmentation algorithms. Preci- sion of liver segmentation based on Bayes classifier is shown in this paper and it is compared with state of the art methods using SLIVER07 dataset.
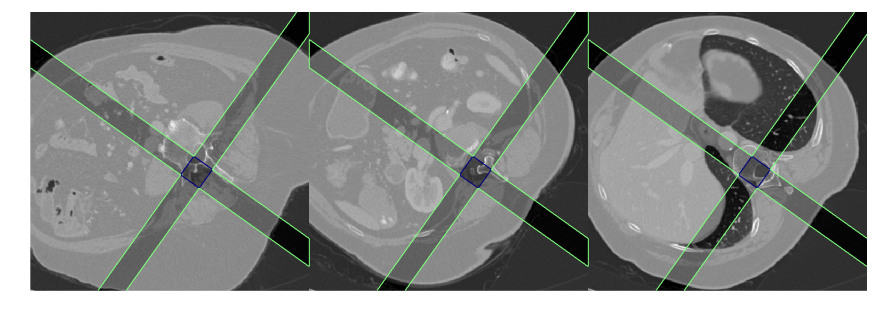
Spine rotation
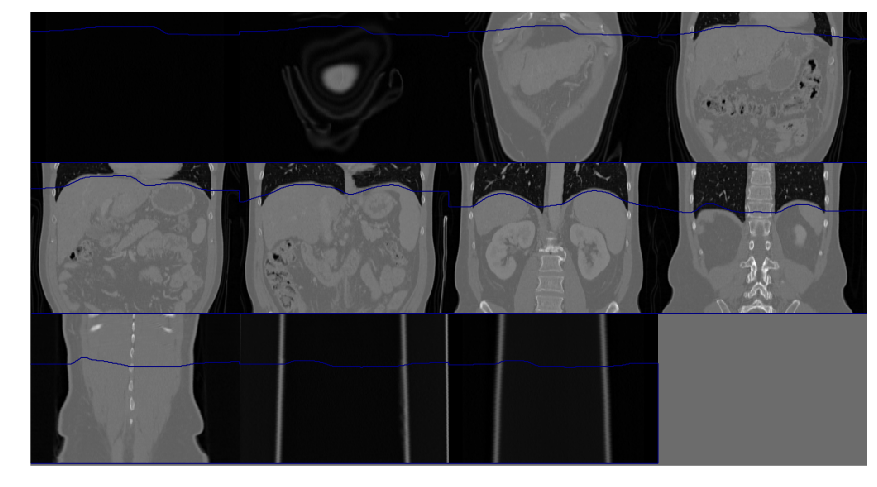
 Diaphragm segmentation
Diaphragm segmentation
Install
conda install -c mjirik bodynavigation
Example
import io3d
import sed3
import bodynavigation
data3d, metadata = io3d.read("dicomdir/")
ss = bodynavigation.body_navigation.BodyNavigation(data3d, metadata["voxelsize_mm"])
seg = ss.get_diaphragm_mask().astype(np.uint8)
sed3.show_slices(data3d, seg*2, slice_step=20, axis=1, flipV=True)
Simple example can be found in examples directory.
Usefull API functions
ss = bodynavigation.body_navigation.BodyNavigation(data3d, metadata["voxelsize_mm"])
dsag = ss.dist_sagittal()
dcor = ss.dist_coronal()
daxi = ss.dist_axial()
ddia = ss.dist_diaphragm()
dsur = ss.dist_to_surface()
dspi = ss.dist_to_spine()
Conda build
conda build . -c conda-forge -c simpleitk -c bioconda --python 3.6