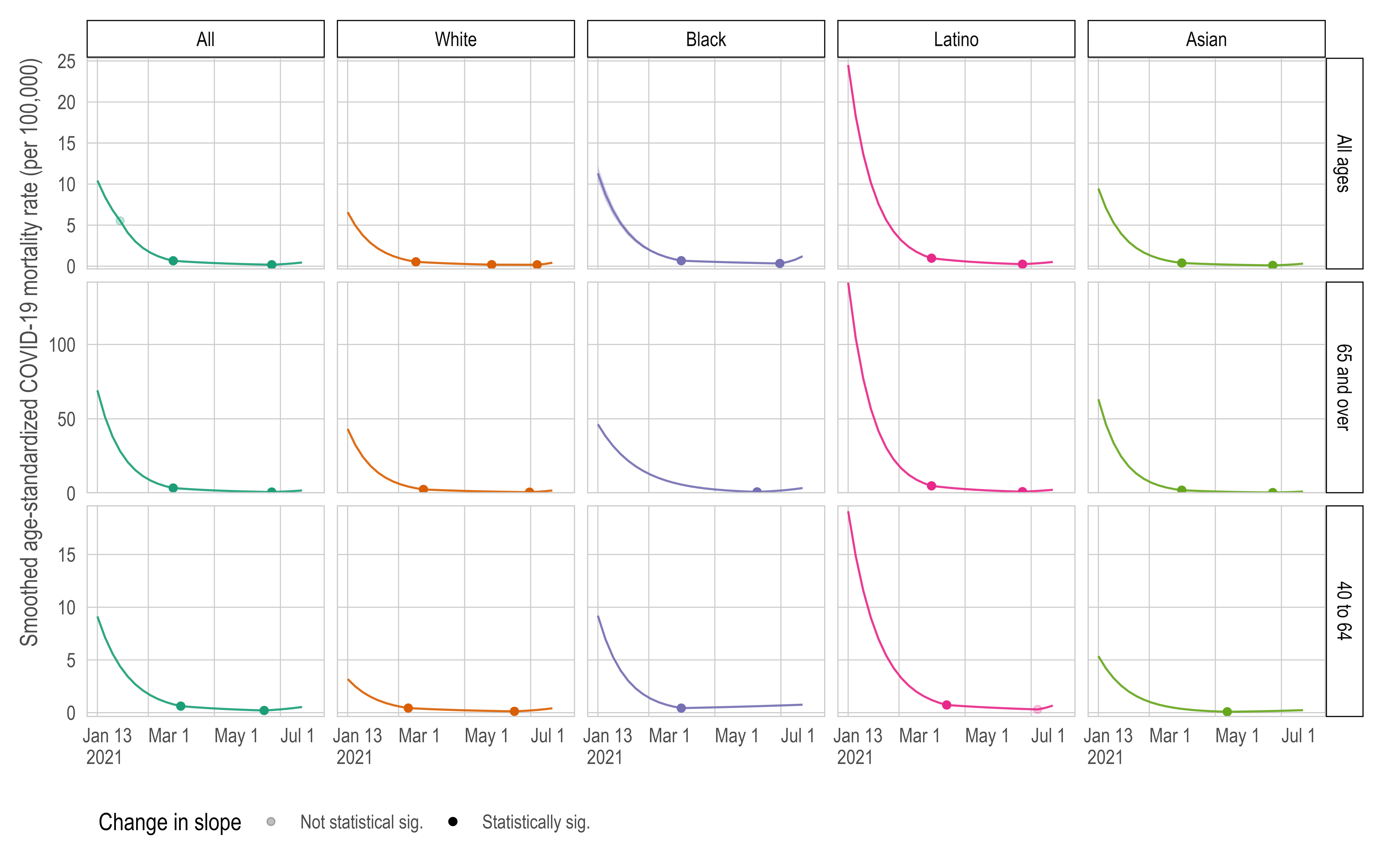
Recent Shifts in Racial/Ethnic Disparities in COVID-19 Mortality in the Vaccination Period in California
This is reproducible code for our recent Journal of General Internal Medicine paper, “Recent Shifts in Racial/Ethnic Disparities in COVID-19 Mortality in the Vaccination Period in California”, which uses restricted-access, decedent-level death certificate data from the California Department of Public Health (CDPH) to systematically examine racial/ethnic disparities in COVID-19 deaths during the vaccination period. The full citation is:
Riley AR†, Kiang MV†, Chen Y-H, Bibbins-Domingo K, & Glymour MM, Recent shifts in racial/ethnic disparities in COVID-19 mortality in the vaccination period in California, Journal of General Internal Medicine (February 2022), doi: 10.1007/s11606-021-07380-6. †First authors contributed equally.
Please submit issues via Github or via email.
In accordance with our data use agreement with the CDPH Vital Statistics, we cannot share individual level data. When possible, we provide aggregated data in cases where there are more than 10 observations. This restriction means this pipeline is not fully reproducible without the restricted-access data.
All analyses are conducted using R, which can be downloaded via
CRAN, and the Joinpoint Regression
Program, which can be downloaded from the National Cancer
Institute.
We also recommend the use of
RStudio when
running R, which will allow users to take advantage of
renv for dependency
management.
The analysis pipeline is divided into three discrete steps.
In Step (1), we clean, subset, munge, and calculate mortality rates
using the raw (restricted-access) data. This results in a working
dataframe that contains the data necessary for the Joinpoint Regression
Program to fit our models of interest. These are held in the 01 to
04 code files.
In Step (2), the joinpoint regressions are fit in an external program
(NCI Joinpoint Regression
Program) and the results
are exported. The ./joinpoint_analyses/age_std_rates.jps file contains
our session information to reproduce our analysis and requires the
./joinpoint_analyses/age_std_rates_long.csv file generated from the
step above (potentially with noise added to small cells).
In Step (3), the resulting (exported) joinpoint files are combined into
a single file for plotting and to create tables. These are code files
05 to 08.
All files should be run sequentially.
- Kirsten
Bibbins-Domingo
(
: @KBibbinsDomingo)
- Yea-Hung Chen
(
: yea-hung |
: @ch272n)
- Maria Glymour
(
: @MariaGlymour)
- Mathew Kiang
(
: mkiang |
: @mathewkiang)
- Alicia
Riley
(
: @aliciacita)