Reproducible code for our paper “Trends in Black and White Opioid Mortality in the United States, 1979-2015” (PDF), which uses multiple cause of death data to examine racial differences in opioid mortality over time. The full citation is:
Alexander MJ, Kiang MV, Barbieri M. Trends in Black and White Opioid Mortality in the United States, 1979–2015. Epidemiology. September 2018. Volume 29, Issue 5, p 707-715. doi: 10.1097/EDE.0000000000000858. Available from: https://journals.lww.com/epidem/Fulltext/2018/09000/Trends_in_Black_and_White_Opioid_Mortality_in_the.16.aspx
Please submit issues via Github or via email.
Please note that the current version of the PDF (8/1/2018) has an error in the abstract. The first sentence of the Results section should read:
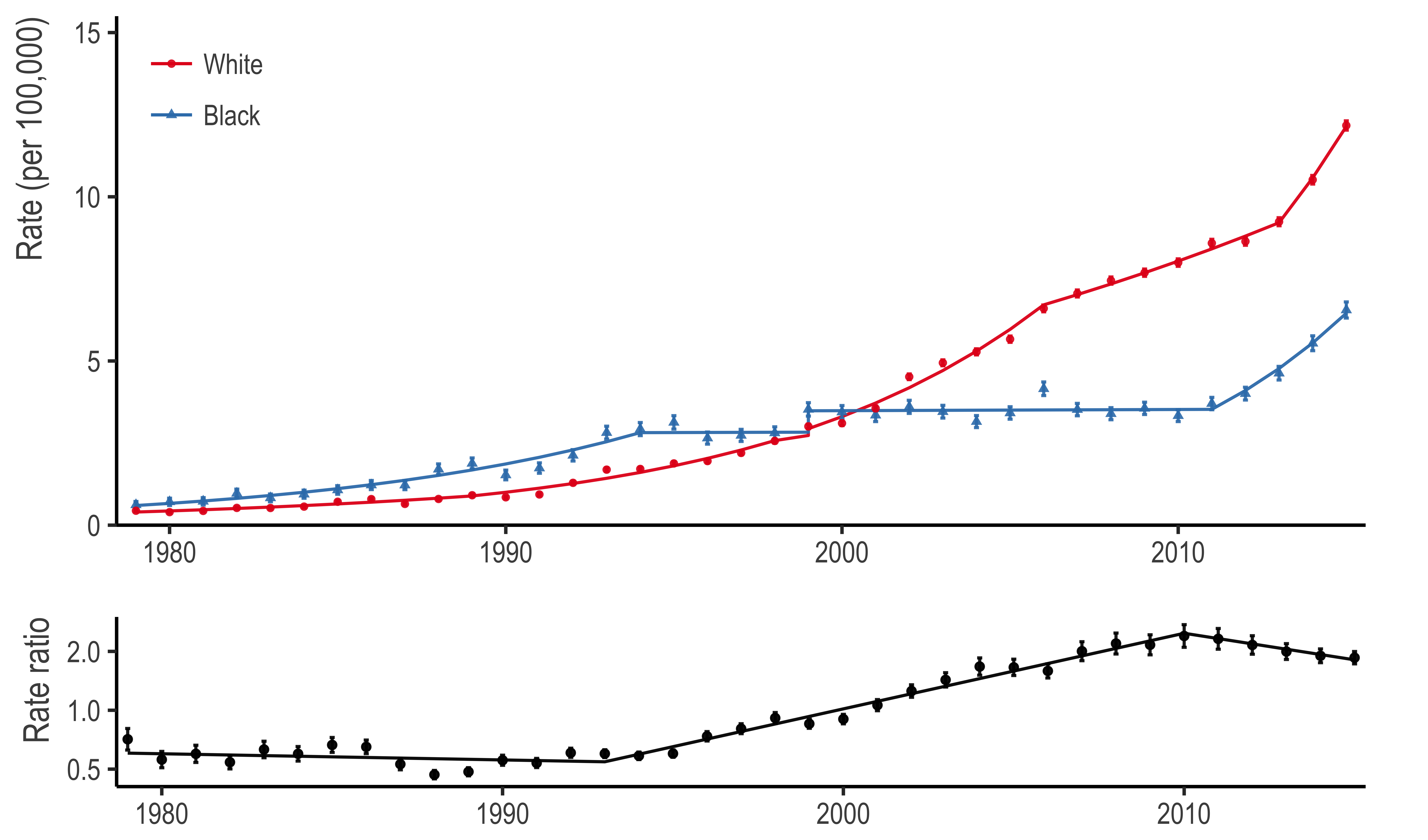
From 1979 to 2015, the long-term trends in opioid-related mortality for US black and white residents went through three successive waves.
We thank Mia Kibel for kindly pointing out this error.
- Submitted: 12/15/2017
- Revisions requested: 1/14/2018
- Revisions submitted: 1/19/2018
- Accepted: 1/22/2018
- Published ahead-of-print online: 5/29/2018
- Publisher’s version available online: 8/1/2018
- Comparing the model fits for 2015 vs 2016 (Mirror): Our data (1979 to 2015) vs adding in the (released-after-submission) 2016 data. (Code.)
- Comparing the model fits for 2015 vs 2017 (Mirror): Our data (1979 to 2015) vs adding in the (released-after-submission) 2017 data. (Code.)
- Counterfactual world: A simple counterfactual analysis where the Black population had the same opioid-related mortality rate as the White population. (Code.)
We use R and the Joinpoint Regression
Program to conduct the
analyses in the paper.[1]
Rcan be downloaded here.- The Joinpoint Regression Program can be downloaded here.
- In addition, we highly recommend the use of RStudio when running
R. RStudio can be downloaded here.
To run this code, you’ll need the following R packages from CRAN:
tidyversehavendoParallelforeachknitrconfigrmarkdownyamldigest
In addition, you’ll need two packages that are not available on CRAN:
- Our package for working with multiple cause of death data,
narcan. - If you want to reproduce our figures exactly, you’ll need
patchwork, but the plots can be generated without it.
You can install these packages manually or by running the
00_install_packages.R script in ./code/. It will not install
packages you already have. It may require interaction (e.g.,
confirmation if a package needs to be compiled). The exact versions we
used can be found in the session_info.txt file.
The analysis pipeline is divided into three parts.
- Part 1: Use
Rto download and munge the data, calculate the rates, and output the rates into a format that the Joinpoint Regression Program can take. - Part 2: Run the joinpoint analyses externally using the output from Part 1.
- Part 3: Ingest the joinpoint regression output and convert it into tables and plots.
Each part has discrete steps and is described in detail below.
The ./config.yml file contains several global parameters for the
analysis pipeline in JSON format. Specifically:
delete_zip_orig: Allows the user to specify if the original MCOD files should be deleted (default:true) or saved (false) after it has been trimmed in Step 1. These files are typically 75 MB per year.delete_trimmed: Allows the user to specify if the smaller MCOD files should be deleted (default:true) or saved (false) after it has been subsetted in the Step 1. These files are typically around 20 MB per year.delete_processed:Allows the user to specify if the uncollapsed MCOD files should be deleted (true) or saved (default:false) after they have been aggregated in Step 3. These files are typically around 15 MB per year. We save them by default to facilitate additional analyses or debugging; however they are not necessary in terms of purely replicating the published analysis.start_yearandend_year: Specify the start (default:1979) and end (default:2015) years of the analysis. Going earlier than 1979 will not work (due to different ICD codes), but as new data gets released, going later than 2015 should work.num_decimals: The number of decimals that should be displayed for Tables 1 and 2num_decimals_supp: The number of decimals that should be displayed for the Supplementary Tablesraw_folder: Specifies where the raw and trimmed MCOD files should be downloaded (default:./raw_data).sav_folder: Specifies where the processed data files (i.e., working data) should be saved (default:./data).output_folder: Specifies where the tables and plots should be saved.proc_in_parallel: Specifies if downloading and processing should be performed in parallel (true) or serially (default:false).
Typically, a user should not need to change any of these parameters;
however, on a computer with sufficient RAM, setting proc_in_parallel
to true should result in significant (linear) speedup. Be warned that
this may result in significant RAM usage (~16 GB of RAM for four
processes) and is not recommended for typical computing environments.
Downloading and cleaning the data on a single processor takes somewhere
in the order of a few hours.
For convenience, the ./01_rerun_step_1.R file will run the following
files for you in a single step. However, it is recommended you open and
inspect each file individually.
- Step 1:
./code/01_download_and_trim_raw_data.R: Downloads data directly from the NBER website and “trims” the dataset by subsetting only to the columns we will use for analysis. In the./config.ymlfile, the user can specify if the raw (untrimmed) data should be kept. The raw data take up approximately 2.9 GB of space (when compressed). The trimmed files take up approximately 900 MB when compressed. When running this process in parallel (that is, setting theproc_in_paralleloption totruein the./config.ymlfile), each process consumes 3.5–4 GB of RAM and the default number of processes is half of the available cores. Make sure your computer is capable of this before setting this option totrue.- Inputs: None
- Outputs:
./raw_data/mortXXXX.dta.zipor./raw_data/mortXXXX.csv.zip(37 files)./raw_data/trimmed_mcod_XXXX.RDS(37 files)
- Step 2:
./code/02_process_trimmed_data.R: This file will perform basic processing on the trimmed multiple cause of death files. Specifically, it will subset to only US residents, clean ICD-9 data issues, convert the age category, add a Hispanic column if necessary for consistency across years, remap the race categories to be consistent across years, and join contributory cause fields for easier string search.- Inputs:
./raw_data/trimmed_mcod_XXXX.RDS(37 files) - Outputs:
./data/cleaned_mcod_XXXX.RDS(37 files)
- Inputs:
- Step 3:
./code/03_flag_opioid_deaths.R: This file will use the underlying cause and contributory cause fields to flag opioid deaths by broad opioid type and when applicable, by ICD-10 type, resulting in our working data set.- Inputs:
./data/cleaned_mcod_XXXX.RDS(37 files) - Outputs:
./data/working_opioid_data.csv
- Inputs:
- Step 4:
./code/04_calculate_mortality_rates.R: This file uses the working data to calculate age-specific and age-standardized mortality rates, by opioid type and race.- Inputs:
./data/working_opioid_data.csv - Outputs:
./data/age_specific_rates.csv./data/age_standardized_rates_wide.csv./data/age_standardized_rates_long.csv
- Inputs:
- Step 5:
./code/05_calculate_opioid_rate_ratio.R: This file uses the working data to calculate the ratio (white/black) of the white and black opioid mortality rates.- Inputs:
./data/working_opioid_data.csv - Outputs:
./data/opioid_rate_ratio.csv
- Inputs:
- Step 6:
./code/06_prepping_data_for_joinpoint.R: The Joinpoint Regression Program needs the data in a specific shape. This file takes in the files calculated from Steps 4 and 5 and reshapes them into the necessary format.- Inputs:
./data/age_standardized_rates_long.csv./data/opioid_rate_ratio.csv
- Outputs:
./joinpoint_analysis/01_opioid_rates_long.csv./joinpoint_analysis/02_opioid_rate_ratio.csv./joinpoint_analysis/03_opioid_rates_by_type.csv./joinpoint_analysis/04_opioid_rates_icd10type.csv
- Inputs:
Part 2 needs to be run outside of R using the Joinpoint Regression
Program (Windows only). To assist with reproducibility, we provide the
original session files (.jps), output files (.jpo), and all the
saved results.
All settings can be inspected by opening the .jps file of
interest[2] and all results can be reviewed by opening the .jpo
file. If you would like to conduct the analysis on your own or change
some program parameters, simply open the .jps file and adjust any
settings.
The ./joinpoint_analysis folder is structured into four analyses,
number 01 to 04. Each one of these analyses contains three files:
the original data (.csv), the joinpoint session file (.jps), and the
saved output (.jpo). In additoin, each analysis also contains a folder
which contains the text-delimited output from the joinpoint regression
program.
For convenience, the ./02_make_plots_and_tables.R file will run the
following files for you in a single step. However, it is recommended you
open and inspect each file individually. Note that we use rmarkdown to
parse and generate the tables. These R scripts below will render the
rmarkdown files into Microsoft Word files; however, if you would like
to see how each file is parsed and sorted, the source of the files are
in ./rmds.
- Step 1:
./code/07_plot_figure1.R: Generate apdfandpngof figure 1.- Inputs: Joinpoint results from
01and02analyses. - Outputs:
fig1_rate_and_ratio.pdffig1_rate_and_ratio.png
- Inputs: Joinpoint results from
- Step 2:
./code/08_plot_figure2.R: Generate apdfandpngof figure 2.- Inputs: Joinpoint results from
03analysis. - Outputs:
fig2_opioid_types.pdffig2_opioid_types.png
- Inputs: Joinpoint results from
- Step 3:
./code/09_plot_figure3.R: Generate apdfandpngof figure 3.- Inputs: Joinpoint results from
04analysis. - Outputs:
fig3_opioid_icd10types.pdffig3_opioid_icd10types.png
- Inputs: Joinpoint results from
- Step 4:
./code/10_generate_tables.R: Generate Microsoft Word documents of Tables 1 and 2.- Inputs:
./rmds/table1_joinpoint_1979_2015.Rmd./rmds/table2_joinpoint_1999_2015.Rmd
- Outputs:
./output/table1_joinpoint_1979_2015.docx./output/table2_joinpoint_1999_2015.docx
- Inputs:
In addition, we provide code that will generate the materials in the
supplement. See ./code/11_plot_efigure1.R and
./code/12_generate_supp_tables.R for more.
Both devtools::session_info() and sessionInfo() output can be found
in the ./session_info.txt file.
- Monica Alexander
(
: mjalexander |
: @monjalexander)
- Magali Barbieri
- Mathew Kiang
(
: mkiang |
: @mathewkiang)
-
We are investigating ways of reproducing the Joinpoint Regression Program using open-source statistical programs, and may update this code in the future.
-
You can also open the output file (
.jpo) and click on “Retrieve Session.”