slendr is an R package which has been primarily designed for simulating spatially-explicit genomic data on real and abstract geographic landscapes. It allows to programmatically and visually encode spatial population boundaries and temporal dynamics across a geographic landscape (leveraging real cartographic data or abstract, user-defined landscapes), and specify population divergences and geneflow events based on an arbitrary graph of demographic history. Additionally, it provides features for simulating large-scale non-spatial population genetics models entirely from R, using both SLiM and msprime as simulation back ends, all without having to leave the convenience of the R environment.
By default, output data is saved in a tree-sequence format, allowing efficient simulations of genome-scale and population-scale data. The R package also provides basic functionality for processing the simulated tree-sequence outputs and for calculating the most frequently used population genetics statistics by implementing an interface to the tree-sequence library tskit via the reticulate package.
By utilizing the flexibility of R with its wealth of libraries for graphics, geospatial analysis and statistics, with the power of the population genetics simulators SLiM and msprime for executing simulations automatically in the background, the slendr R package makes it possible to write entire simulation and analytic pipelines without the need to leave the R environment.
You can open an RStudio session and test examples from the vignettes directly in your web browser by clicking this button (no installation is needed):
In case the RStudio instance appears to be starting very slowly, please be patient. The binder cloud server can sometimes take a minute to load up. Sometimes it even crashes completely. If that happens, try reloading the page - this will restart the binder session.
Once you get a browser-based RStudio session, you can navigate to the vignettes/ directory and test the examples on your own!
This software is still under development! We have been making steady progress towards the first beta version, but the package still has some way to go before being production ready. Please update the slendr installation by running devtools::install_github("bodkan/slendr") regularly.
That said, if you would like to learn more, or if you're feeling brave and would like to test the package yourself, take a look at some of our tutorial vignettes (either the main tutorial or other vignettes available under the "Articles" menu on the project website).
If you would like to stay updated with the developments:
-
Click on the "Watch" button on the project's Github website.
-
Follow me on Twitter. I might post some updates once the software is a bit more ready.
For installation instructions, please take a look at the installation section of the main tutorial. Note that in order to simulate data, you will need the most recent version of the SLiM software (version 3.6 or later). Similarly, if you would like to process tree-sequence data generated by slendr & SLiM from R, you will need a working Python environment with the modules tskit and pyslim (see the relevant vignette for more detail).
The main dependency of slendr is the R package sf, which it uses for manipulation of geospatial data. In turn, sf depends on several external pieces of software which sometimes need to be installed. That said, if you can successfully install sf by running install.packages("sf") you will be good to go. In case you run into trouble, following sf's installation guidelines has so far always been a success.
Here is a small demonstration of what slendr is designed to do. We want to simulate spatiotemporal data representing the history of modern humans in Eurasia after the Out of Africa migration. This example will be quite brief, for more details, please see the tutorial vignette.
First, we define the spatial context of the simulation. This will be the entire "world" which will be occupied by populations in our model. Note that in the world definition, we are explicitly stating which projected Coordinate Reference System (CRS) will be used to represent landscape features, distances in kilometers, etc.
library(slendr)
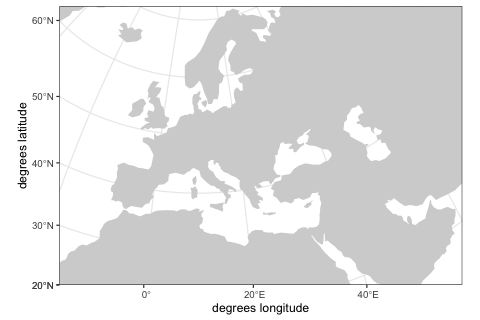
map <- world(
xrange = c(-15, 60), # min-max longitude
yrange = c(20, 65), # min-max latitude
crs = "EPSG:3035" # real projected CRS used internally
)We can visualize the defined world map using the generic function plot provided by the package.
plot(map)Although in this example we use a real Earth landscape, the map can be completely abstract (either blank or with user-defined landscape features such as continents, islands, corridors and barriers).
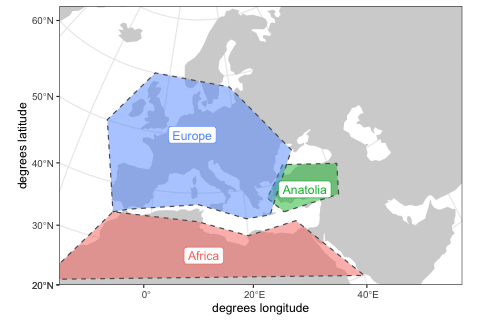
In order to make building of population boundaries easier, we can define smaller regions on the map using the function region.
Note all coordinates of are specified in the geographic coordinate system (degrees longitude and latitude), but are internally represented in a projected CRS. This makes it easier to define spatial features simply by reading the coordinates from any regular map but makes simulations more accurate (distances and shapes are not distorted because we can use a CRS tailored to the region of the world we are working with).
africa <- region(
"Africa", map,
polygon = list(c(-18, 20), c(40, 20), c(30, 33),
c(20, 32), c(10, 35), c(-8, 35))
)
europe <- region(
"Europe", map,
polygon = list(
c(-8, 35), c(-5, 36), c(10, 38), c(20, 35), c(25, 35),
c(33, 45), c(20, 58), c(-5, 60), c(-15, 50)
)
)
anatolia <- region(
"Anatolia", map,
polygon = list(c(28, 35), c(40, 35), c(42, 40),
c(30, 43), c(27, 40), c(25, 38))
)Again, we can use the generic plot function to visualize the objects:
plot(africa, europe, anatolia)The most important function in the package is population, which is used to define names, split times, sizes and spatial ranges of populations. Here, we specify times in years before the present, distances in kilometers. If this makes more sense for your models, times can also be given in a forward direction.
You will also note functions such as move or expand which are designed to take a slendr population object and change its spatial dynamics.
Note that in order to make this example executable on a normal local machine, we deliberately decreased the sizes of all populations.
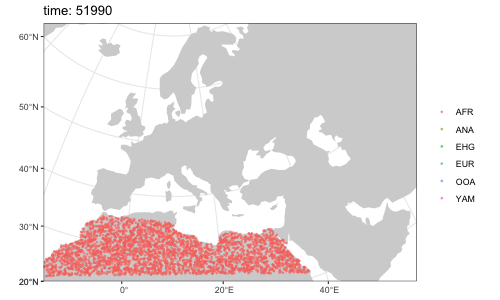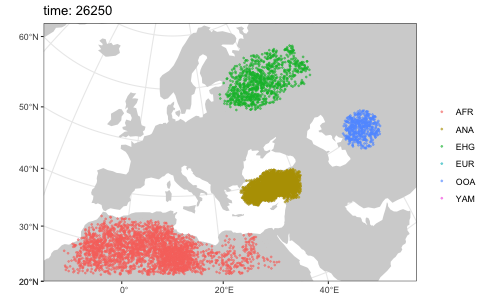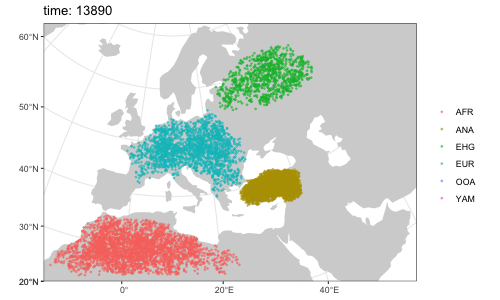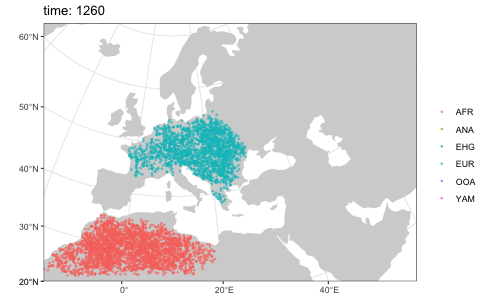
afr <- population( # African ancestral population
"AFR", parent = "ancestor", time = 52000, N = 3000,
map = map, polygon = africa
)
ooa <- population( # population of the first migrants out of Africa
"OOA", parent = afr, time = 51000, N = 500, remove = 25000,
center = c(33, 30), radius = 400e3
) %>%
move(
trajectory = list(c(40, 30), c(50, 30), c(60, 40)),
start = 50000, end = 40000, snapshots = 20
)
ehg <- population( # Eastern hunter-gatherers
"EHG", parent = ooa, time = 28000, N = 1000, remove = 6000,
polygon = list(
c(26, 55), c(38, 53), c(48, 53), c(60, 53),
c(60, 60), c(48, 63), c(38, 63), c(26, 60))
)
eur <- population( # European population
name = "EUR", parent = ehg, time = 25000, N = 2000,
polygon = europe
)
ana <- population( # Anatolian farmers
name = "ANA", time = 28000, N = 3000, parent = ooa, remove = 4000,
center = c(34, 38), radius = 500e3, polygon = anatolia
) %>%
expand( # expand the range by 2.500 km
by = 2500e3, start = 10000, end = 7000,
polygon = join(europe, anatolia), snapshots = 20
)
yam <- population( # Yamnaya steppe population
name = "YAM", time = 7000, N = 500, parent = ehg, remove = 2500,
polygon = list(c(26, 50), c(38, 49), c(48, 50),
c(48, 56), c(38, 59), c(26, 56))
) %>%
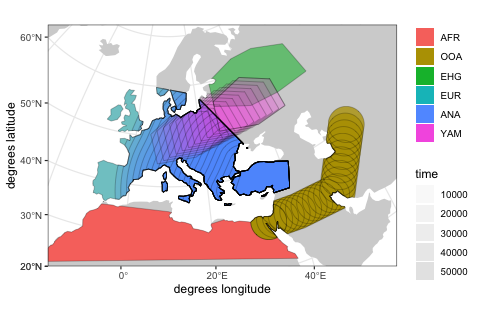
move(trajectory = list(c(15, 50)), start = 5000, end = 3000, snapshots = 10)We can use the function plot again, but we get a warning informing us that plotting complex model dynamics over time on a single map is not a good idea. Below, we show a better way to do this using a built-in interactive R shiny app.
plot(afr, ooa, ehg, eur, ana, yam)By default, overlapping populations in SLiM do not mix. In order to schedule an geneflow event between two populations, we can use the function geneflow. If we want to specify multiple such events at once, we can collect them in a simple R list:
gf <- list(
geneflow(from = ana, to = yam, rate = 0.5, start = 6500, end = 6400, overlap = FALSE),
geneflow(from = ana, to = eur, rate = 0.5, start = 8000, end = 6000),
geneflow(from = yam, to = eur, rate = 0.75, start = 4000, end = 3000)
)model <- compile(
populations = list(afr, ooa, ehg, eur, ana, yam), # populations defined above
geneflow = gf, # geneflow events defined above
generation_time = 30,
resolution = 10e3, # resolution in meters per pixel
competition_dist = 130e3, mate_dist = 100e3, # spatial interaction in SLiM
dispersal_dist = 70e3, # how far will offspring end up from their parents
path = file.path(tempdir(), "readme-model"), overwrite = TRUE
)Compiled model is kept as an R object which can be passed to different functions, most importantly the slim() function shown below.
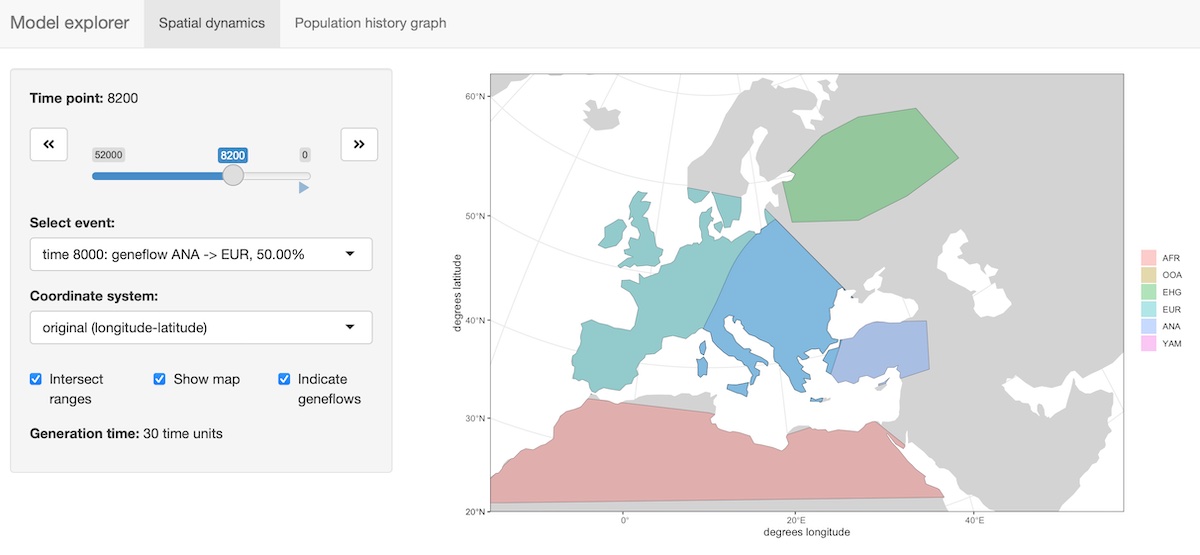
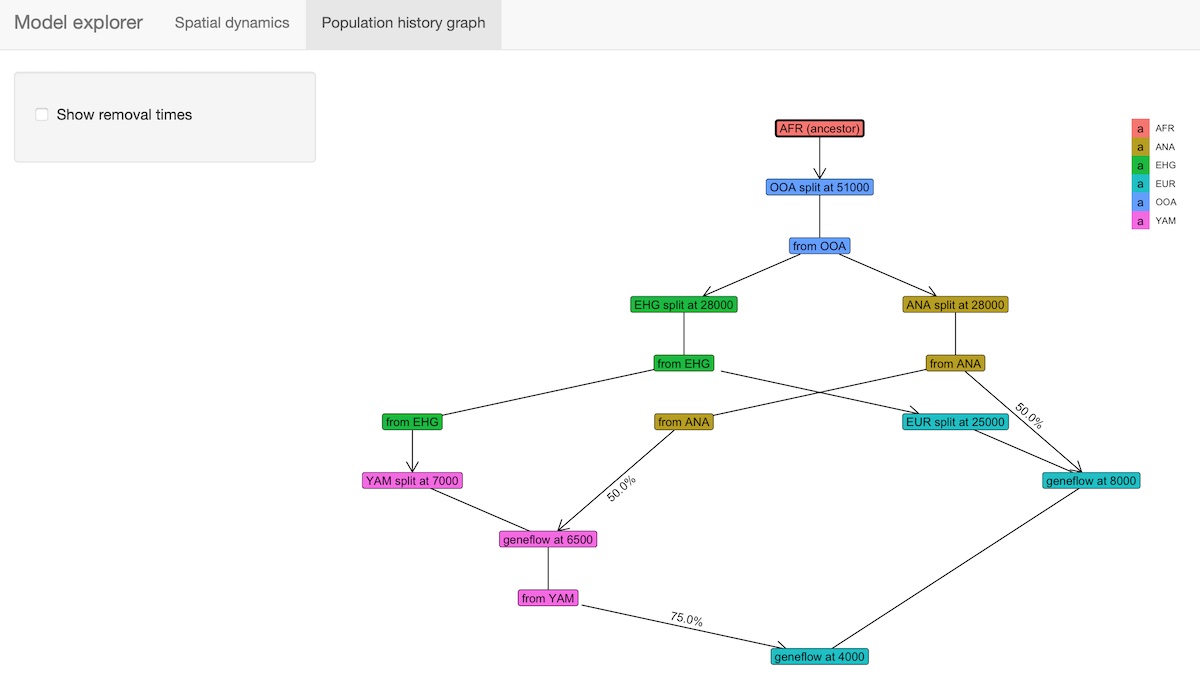
The package provides an R shiny-based browser app explore() for checking the model dynamics interactively and visually. For more complex models, this is much better than static spatial plots such as the one we showed in step 2 above:
explore(model)The function has two modes:
a) Plotting spatial map dynamics:
b) Displaying the demographic history graph (splits and geneflow events) embedded in the specified model:
Finally, we can execute the compiled model in SLiM. Here we run the simulation in a batch mode, but we could also run it in SLiMgui by setting method = "gui".
The slim function generates a complete SLiM script tailored to run the spatial model we defined above. This saves you, the user, a tremendous amount of time.
slim(
model,
sequence_length = 1, recombination_rate = 0, # simulate only a single locus
save_locations = TRUE, # save the location of everyone who ever lived
method = "batch", # change to "gui" to execute the model in SLiMgui
random_seed = 314159
)As specified, the SLiM run will save ancestry proportions in each population over time as well as the location of every individual who ever lived.
We can use the saved locations of every individual that lived throughout the course of the simulation to generate a simple GIF animation (please note that the animation has been significantly sped up to decrease the GIF file size):
animate(model = model, steps = 50, width = 500, height = 300)Note that it is possible to simulate population splits and geneflows both by "physically" moving individuals of a population from one destination to the next across space but it is also possible to do this more abstractly (in instantaneous "jumps") in situations where this is more appropriate or where simulating accurate movement is not necessary.
In this case, the only population movement we explicitly encoded was the split and migration of the OOA population from Africans and the expansion of Anatolians. All the other populations simply popped up on a map, without explicitly migrating there. Similarly, the geneflow from Anatolians into the Yamnaya specified in step 4. did not require a spatial overlap between the two populations, but the other two geneflow events did.
The example above provides only a very brief overview of the functionality of the slendr package. There is much more to slendr than what we demonstrated here. For instance:
-
You can tweak parameters influencing dispersal dynamics (how "clumpy" populations are, how far can offspring migrate from their parents, etc.) and define how these should change over time. For instance, you can see that in the animation above, the African population forms a single "blob" that really isn't spread out across its entire population range. Tweaking the dispersal parameters as show this vignette helps avoid that.
-
You can use slendr to program non-spatial models, which means that any concievable demographic model can be simulated with only a few lines of R code and, for instance, plugged into an Approximate Bayesian Computation pipeline or other use any other R package for downstream analysis in the same R script. You can find more in this vignette. Because SLiM simulations can be often quite slow compared to their coalescent counterperts, we also provide functionality allowing to simulate slendr models (without any change!) using a built-in msprime back end script. See this vignette for a tutorial on how this works.
-
You can build complex spatial models which are still abstract (not assuming any real geographic location), including traditional simulations of demes in a lattice structure. A complete example is shown this vignette.
-
Because slendr & SLiM save data in a tree-sequence file format, thanks to the R package reticulate for interfacing with Python code, we have the incredible power of tskit and pyslim for manipulating tree-sequence data right at our fingertips, all within the convenient environment of R. An extended example can be found in this vignette.
-
For spatially explicit population models, the slendr package automatically converts the simulated output data to a format which makes it possible to analyse it with many available R packages for geospatial data analysis. A brief description of this functionality can be found in this vignette.