Introduction
The symbolicpedprob package implements symbolic evaluation of pedigree likelihoods. So far, the likelihood can only be evaluated for a single marker without the use of subpopulation correction or mutation modelling.
The main function of the package is the sLikelihood() function. The
aim is to mimic the likelihood() function of the pedprobr package
which implements numeric evaluation of pedigree likelihoods.
Getting started
library(symbolicpedprob)
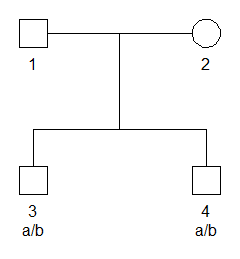
library(pedtools)To set up a simple example, we first use pedtools utilities to
create a pedigree where two brothers are genotyped with a single SNP
marker. The marker has alleles a and b, and both brothers are
heterozygous a/b.
# Pedigree with SNP marker
x = nuclearPed(nch = 2) |>
addMarker(geno = c(NA, NA, "a/b", "a/b"))
# Plot with genotypes
plot(x, marker = 1)The pedigree likelihood, i.e., the probability of the genotypes given the pedigree, is obtained as follows:
sLikelihood(x)
#> [1] "a^2*b^2+0.5*a^2*b+0.5*a*b^2+0.5*a*b"