IN PROGRESS!
{reactibble} implements reactive columns in data frames.
We build them using the reactibble() function, and define reactive
columns using formulas.
library(reactibble)
rt <- reactibble(
height = c(1.50, 1.72),
weight = c(70, 82),
bmi = ~ weight / height^2
)As you can see, the reactive columns are displayed in a different color,
and the type of the column is prefixed with "~".
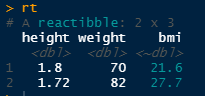
Then whenever we change a value, the reactive column is updated.
rt$height[1] <- 1.80It is reminescent database views, excel calculated columns or reactive
DataTables in {shiny}.
The package provides functions and methods to modify the data and ease memoisation of operations.
Install with:
remotes::install_github("moodymudskipper/reactibble")You can add or modify columns using {dplyr} functions mutate() and
transmute(), use a formula if you wish to add a new reactive column.
Building on our example above :
library(dplyr, warn.conflicts = FALSE)
rt <- mutate(rt, height_cm = ~ height * 100)
rt
#> # A reactibble: 2 x 4
#> height weight bmi height_cm
#> <dbl> <dbl> <~dbl> <~dbl>
#> 1 1.8 70 21.6 180
#> 2 1.72 82 27.7 172To add rows we can use the functions rt_bind_rows() or rt_add_row(),
counterpart to dplyr::bind_rows() and tibble::add_row() which
unfortunately don’t work reliably on "reactibble" objects.
rt <- rt_add_row(rt, height = 1.64, weight = 68)
rt
#> # A reactibble: 3 x 4
#> height weight bmi height_cm
#> <dbl> <dbl> <~dbl> <~dbl>
#> 1 1.8 70 21.6 180
#> 2 1.72 82 27.7 172
#> 3 1.64 68 25.3 164To get a static object (still of class reactibble), call materialize()
materialize(rt)
#> # A reactibble: 3 x 4
#> height weight bmi height_cm
#> <dbl> <dbl> <dbl> <dbl>
#> 1 1.8 70 21.6 180
#> 2 1.72 82 27.7 172
#> 3 1.64 68 25.3 164We implemented methods for {dplyr}’s join functions so that they
should work seamlessly on "reactibble" reactibble objects, provided
the first argument is a "reactibble".
Renaming, selecting, dropping columns, slicing, will work seamlessly with the method of your choice. In case of renaming the formula of the reactive columns will adapt to the new names, and if a necessary column is dropped an explicit error will be triggered.
Some functions outside of this package will work well natively on
{reactibble}, such as base::split(), others will warn, such as
base::transform(), while others might unfortunately strip off the
class or attributes silently, or put the reactibble out of sync.
We hope through time to provide methods for most generics posing
problems, and rt_* alternatives for other problematic functions, if
you want to help,
The columns of a "reactibble" object are recomputed anytime the object
is modified. In some cases this can be costly in terms of resources, we
can deal with this either by memoising the columns (next section) or
enabling manual refresh (present section).
We can disable the autorefresh, do the transformation we need with the latest computed values, and refresh once we’re done.
options(reactibble.autorefresh = FALSE)
rt <- mutate(rt, weight_g = ~weight * 1000)
# not yet refreshed
rt
#> # A reactibble: 3 x 5
#> height weight bmi height_cm weight_g
#> <dbl> <dbl> <~dbl> <~dbl> <~lgl>
#> 1 1.8 70 21.6 180 NA
#> 2 1.72 82 27.7 172 NA
#> 3 1.64 68 25.3 164 NA
# now refreshed
rt <- refresh(rt)
rt
#> # A reactibble: 3 x 5
#> height weight bmi height_cm weight_g
#> <dbl> <dbl> <~dbl> <~dbl> <~dbl>
#> 1 1.8 70 21.6 180 70000
#> 2 1.72 82 27.7 172 82000
#> 3 1.64 68 25.3 164 68000
# go back to default
options(reactibble.autorefresh = TRUE)The other, arguably more convenient way to deal with redundant
computations is to memoise them. For this we implemented a handy feature
that wraps the {memoise} package which .
Wrap the function that you want to memoise in M() and whenever its fed
the same input when refreshing the column, it will retrieve the result
in memory.
See example below where we memoise respectively the full column and rowwise results.
slow_mean <- function(...) {
Sys.sleep(1)
mean(c(...))
}
# the function is run the first time, taking one second
system.time(
rt <- reactibble(a=1:3, b = ~ M(slow_mean)(a))
)
#> user system elapsed
#> 0.00 0.02 1.05
rt
#> # A reactibble: 3 x 2
#> a b
#> <int> <~dbl>
#> 1 1 2
#> 2 2 2
#> 3 3 2
# If we transform the data without affecting b, we use results stored in
# memory, saving resources
system.time(
rt <- mutate(rt, c = ~ b * 2)
)
#> user system elapsed
#> 0.05 0.00 0.04We can memoise rowwise computations that wrapping the call in an apply
function (lapply(), sapply(), mapply(), Map(), or functions from
the purrr::map*() family)
# the function is executed 3 times, takes 3 seconds
system.time(
rt <- reactibble(a=1:3, b = 2:4, c = ~ mapply(M(slow_mean),a, b))
)
#> user system elapsed
#> 0.02 0.00 3.14
rt
#> # A reactibble: 3 x 3
#> a b c
#> <int> <int> <~dbl>
#> 1 1 2 1.5
#> 2 2 3 2.5
#> 3 3 4 3.5
# if we add a single row, it will take only one more second to update
system.time(
# careful not to change the type from integer to double!
rt <- rt_add_row(rt, a=4L, b = 5L)
)
#> user system elapsed
#> 0.00 0.00 1.07
rt
#> # A reactibble: 4 x 3
#> a b c
#> <int> <int> <~dbl>
#> 1 1 2 1.5
#> 2 2 3 2.5
#> 3 3 4 3.5
#> 4 4 5 4.5