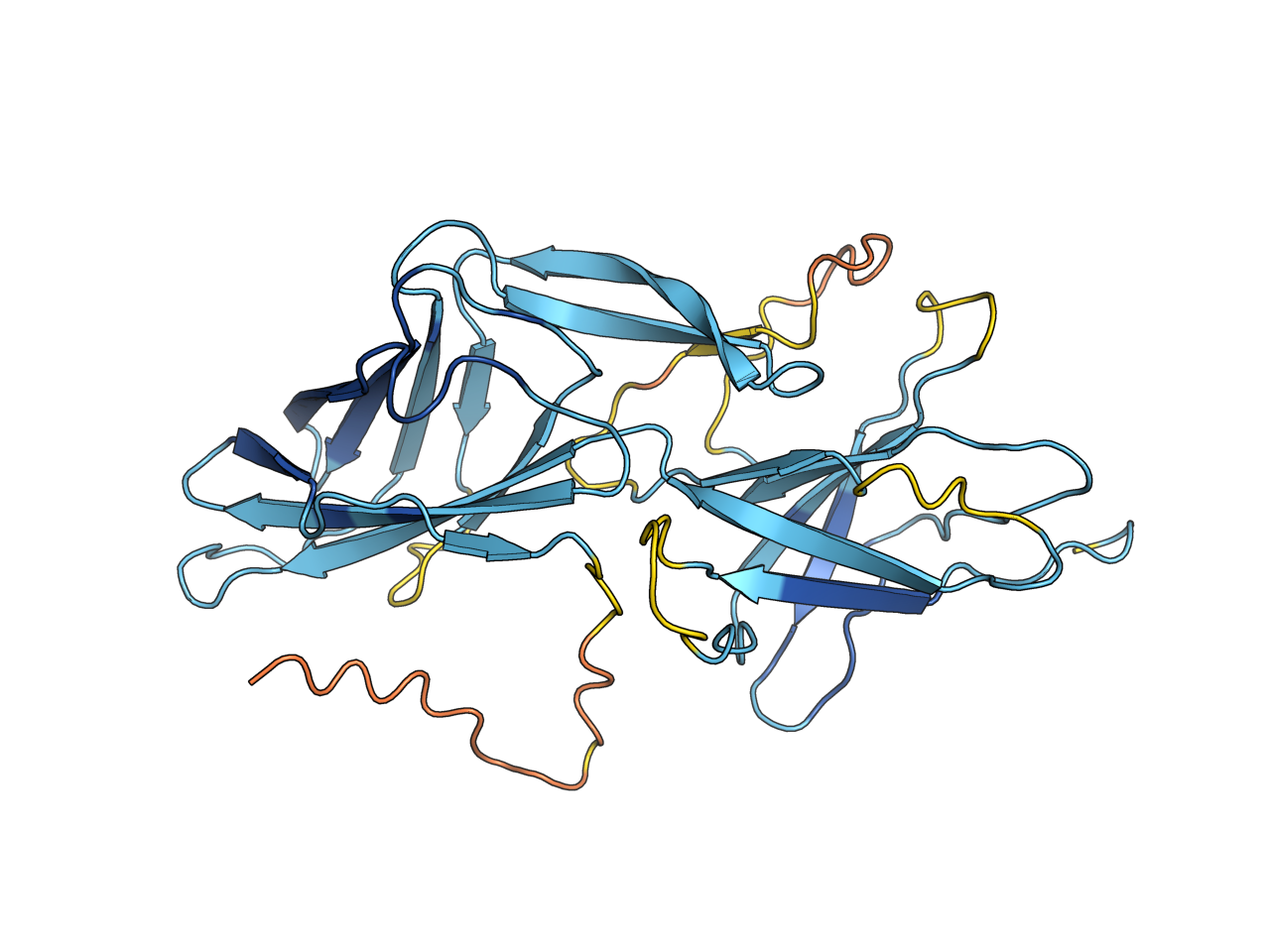
Fold your protein in PyMOL!
Inspired by ColabFold by Sergey O.
Visualization inspired by pymol-color-alphafold.
Thanks to ESMFold by Meta and the API.
Tested under macOS Monterey Version 12.5.1, Python 3.7.12.
Open an issue if ran into any errors.
03Dec2022: Add `dms`, `singlemut`, and `webapps`. `pymolfold` allow sequence length up to 700aa.
26Nov2022: ProteinMPNN is now integrated to design proteins.
15Nov2022: We now provide an unofficial API to support user defined recycle number and allow sequence length up to 500aa!
conda install -c conda-forge pymol-open-sourcerun https://raw.githubusercontent.com/JinyuanSun/PymolFold/main/predict_structure.py
# for user still using python2, it is also py3 compatible, only esmfold supports.
run https://raw.githubusercontent.com/JinyuanSun/PymolFold/py27/predict_structure.py
# try the command below in China mainland, the mirror will be delayed if modifications were just made, download the file to your computer and install it is always a good idea:
run https://raw.staticdn.net/JinyuanSun/PymolFold/main/predict_structure.pywebapp avaiable at here, in case someone struggles with using PyMOL.
Also, check META's web app
The color_plddt command also returns pymol selection object of different confidence levels. The color scheme is now compatible with plddt in range (0, 1) and (0, 100) only if they are consistent in your selection.
esmfold GENGEIPLEIRATTGAEVDTRAVTAVEMTEGTLGIFRLPEEDYTALENFRYNRVAGENWKPASTVIYVGGTYARLCAYAPYNSVEFKNSSLKTEAGLTMQTYAAEKDMRFAVSGGDEVWKKTPTANFELKRAYARLVLSVVRDATYPNTCKITKAKIEAFTGNIITANTVDISTGTEGSGTQTPQYIHTVTTGLKDGFAIGLPQQTFSGGVVLTLTVDGMEYSVTIPANKLSTFVRGTKYIVSLAVKGGKLTLMSDKILIDKDWAEVQTGTGGSGDDYDTSFN, test
color_plddt
orient
ray 1280, 960, async=1pymolfold GENGEIPLEIRATTGAEVDTRAVTAVEMTEGTLGIFRLPEEDYTALENFRYNRVAGENWKPASTVIYVGGTYARLCAYAPYNSVEFKNSSLKTEAGLTMQTYAAEKDMRFAVSGGDEVWKKTPTANFELKRAYARLVLSVVRDATYPNTCKITKAKIEAFTGNIITANTVDISTGTEGSGTQTPQYIHTVTTGLKDGFAIGLPQQTFSGGVVLTLTVDGMEYSVTIPANKLSTFVRGTKYIVSLAVKGGKLTLMSDKILIDKDWAEVQTGTGGSGDDYDTSFN, 4, test
color_plddt
orient
ray 1280, 960, async=1Thanks to ColabDeisgn by Sergey O.
cpd for sequence generation Webapp
Use cpd to design seqeunces will fold into the target structure:
# commands
fetch 1pga.A
cpd 1pga.A
# output looks like:
# >des_0,score=0.72317,seqid=0.6607
# PTYKLIINGKKIKGEISVEAPDAKTAEKIFKNYAKENGVNGKWTYDESTKTFTIEE
# >des_1,score=0.73929,seqid=0.6250
# PTYTLVVNGKKIKGTRSVEAPNAAVAEKIFKQWAKENGVNGTWTYDASTKTFTVTE
# >des_2,score=0.72401,seqid=0.6429
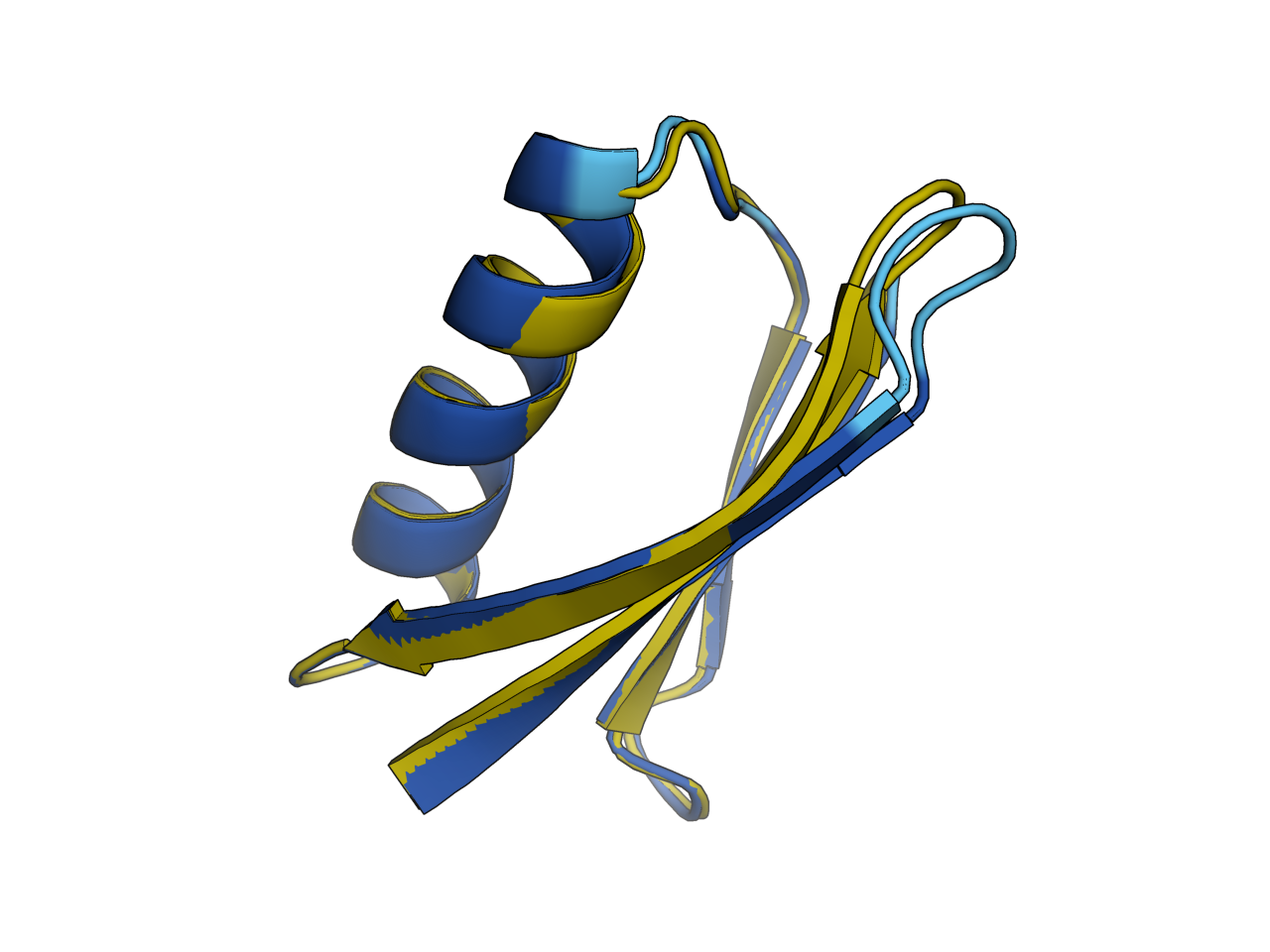
# PTYTLKINGKKIKGEISVEAPNAEEAEKIFKQYAKDHGVNGKWTYDASTKTFTVTEUsing esmfold to examin the des_0:
# commands
esmfold PTYKLIINGKKIKGEISVEAPDAKTAEKIFKNYAKENGVNGKWTYDESTKTFTIEE, 1pga_des0
super 1pga_des0, 1pga.A
color_plddt 1pga_des0singlemut for scoring a signle mutation Webapp
# commands
fetch 1pga.A
singlemut 1pga.A, A, 26, F
# output maybe (not deterministic):
# ================================
# mutation: A_26_F, score: -0.0877
# ================================dms for in silico deep mutational scan Webapp
# commands
fetch 1pga.A
select resi 1-10
dms sele
# this might took ~1 min, be pacient ; )
# output:
# Results save to '/pat/to/working/dir/dms_results.csv'@article{lin2022language,
title={Language models of protein sequences at the scale of evolution enable accurate structure prediction},
author={Lin, Zeming and Akin, Halil and Rao, Roshan and Hie, Brian and Zhu, Zhongkai and Lu, Wenting and dos Santos Costa, Allan and Fazel-Zarandi, Maryam and Sercu, Tom and Candido, Sal and others},
journal={bioRxiv},
year={2022},
publisher={Cold Spring Harbor Laboratory}
}
@article{
doi:10.1126/science.add2187,
author = {J. Dauparas and I. Anishchenko and N. Bennett and H. Bai and R. J. Ragotte and L. F. Milles and B. I. M. Wicky and A. Courbet and R. J. de Haas and N. Bethel and P. J. Y. Leung and T. F. Huddy and S. Pellock and D. Tischer and F. Chan and B. Koepnick and H. Nguyen and A. Kang and B. Sankaran and A. K. Bera and N. P. King and D. Baker },
title = {Robust deep learning-based protein sequence design using ProteinMPNN},
journal = {Science},
volume = {378},
number = {6615},
pages = {49-56},
year = {2022},
doi = {10.1126/science.add2187},
URL = {https://www.science.org/doi/abs/10.1126/science.add2187}
}
PyMOL is a trademark of Schrodinger, LLC.