How to handle missing or incomplete data
One subject that often crops up is how to handle missing or incomplete data.
I decided to try this tutorial to get some background on the issue. The general approach will be as follows:
- Describe the data
- Check for missing values
- Fill in any missing values
- Compare the filled-in values with the original values
Following on from my ML with SciPy exercise, I make sure to carefully examine the structure of the data first!
The table of contents is as follows:
This is a long-standing issue. If a sensitive or troublesome field is left as optional, it will tend to be either: left blank, or else populated with values such as N/A (meaning possibly "Not Applicable" or "Not Available"). So, using SICs (Sales Industry Codes - which are generally three digits) as an example, if this field is made mandatory - and validated for being numeric - the easy data entry options will tend to be either "000" or "999" (although other options for "unknown" Sales Industry Codes are of course possible). But none of these values make for good data analysis.
[The essential problem is that data entry personnel generally lack both the training and the data to correctly determine the missing fields. Plus they are generally paid by volume, so it is not really in their best interests to spend a lot of time on their data-entry problems.]
We will use the Pima Indians Diabetes dataset.
As it no longer seems to be available, we will use the tutorial author's version.
This data is known to have missing values. It consists of:
- Number of times pregnant
- Plasma glucose concentration a 2 hours in an oral glucose tolerance test
- Diastolic blood pressure (mm Hg)
- Triceps skinfold thickness (mm)
- 2-Hour serum insulin (mu U/ml)
- Body mass index (weight in kg/(height in m)^2)
- Diabetes pedigree function
- Age (years)
- Class variable (0 or 1)
This looks as follows:
$ python missing_data.py
Rows, columns = (768, 9)
The first 20 observations
-------------------------
0 1 2 3 4 5 6 7 8
0 6 148 72 35 0 33.6 0.627 50 1
1 1 85 66 29 0 26.6 0.351 31 0
2 8 183 64 0 0 23.3 0.672 32 1
3 1 89 66 23 94 28.1 0.167 21 0
4 0 137 40 35 168 43.1 2.288 33 1
5 5 116 74 0 0 25.6 0.201 30 0
6 3 78 50 32 88 31.0 0.248 26 1
7 10 115 0 0 0 35.3 0.134 29 0
8 2 197 70 45 543 30.5 0.158 53 1
9 8 125 96 0 0 0.0 0.232 54 1
10 4 110 92 0 0 37.6 0.191 30 0
11 10 168 74 0 0 38.0 0.537 34 1
12 10 139 80 0 0 27.1 1.441 57 0
13 1 189 60 23 846 30.1 0.398 59 1
14 5 166 72 19 175 25.8 0.587 51 1
15 7 100 0 0 0 30.0 0.484 32 1
16 0 118 84 47 230 45.8 0.551 31 1
17 7 107 74 0 0 29.6 0.254 31 1
18 1 103 30 38 83 43.3 0.183 33 0
19 1 115 70 30 96 34.6 0.529 32 1Examining the first 20 observations, we can see zeroes (but no troublesome "99" or "999" values - perhaps medical personnel are closer to the data) in a number of columns. It is only reasonable that there should be zeroes in the first and last columns. So we will check for zeroes in all of the other columns:
Number of zero values
---------------------
1 5
2 35
3 227
4 374
5 11
6 0
7 0
dtype: int64It looks like the only problems areas are columns 1, 2, 3, 4 and 5.
According to the tutorial, it is standard practice in Python (specifically Pandas, NumPy and Scikit-Learn) to mark missing values as NaN.
Firstly, check for missing values using the Pandas isnull function before doing any data munging:
Number of missing fields (original)
-----------------------------------
0 0
1 0
2 0
3 0
4 0
5 0
6 0
7 0
8 0
dtype: int64
Statistics (original)
---------------------
0 1 2 3 4 5 \
count 768.000000 768.000000 768.000000 768.000000 768.000000 768.000000
mean 3.845052 120.894531 69.105469 20.536458 79.799479 31.992578
std 3.369578 31.972618 19.355807 15.952218 115.244002 7.884160
min 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000
25% 1.000000 99.000000 62.000000 0.000000 0.000000 27.300000
50% 3.000000 117.000000 72.000000 23.000000 30.500000 32.000000
75% 6.000000 140.250000 80.000000 32.000000 127.250000 36.600000
max 17.000000 199.000000 122.000000 99.000000 846.000000 67.100000
6 7 8
count 768.000000 768.000000 768.000000
mean 0.471876 33.240885 0.348958
std 0.331329 11.760232 0.476951
min 0.078000 21.000000 0.000000
25% 0.243750 24.000000 0.000000
50% 0.372500 29.000000 0.000000
75% 0.626250 41.000000 1.000000
max 2.420000 81.000000 1.000000 Now we will use the Pandas replace function to replace our troublesome zero values with NaN.
And check again for zero (missing) values:
Number of missing fields (zero fields flagged as NaN)
-----------------------------------------------------
0 0
1 5
2 35
3 227
4 374
5 11
6 0
7 0
8 0
dtype: int64And columns 1, 2, 3, 4 and 5 have missing values.
Lets get the stats for the columns we will be filling:
Statistics (pre-fill)
---------------------
1 2 3 4 5
count 763.000000 733.000000 541.000000 394.000000 757.000000
mean 121.686763 72.405184 29.153420 155.548223 32.457464
std 30.535641 12.382158 10.476982 118.775855 6.924988
min 44.000000 24.000000 7.000000 14.000000 18.200000
25% 99.000000 64.000000 22.000000 76.250000 27.500000
50% 117.000000 72.000000 29.000000 125.000000 32.300000
75% 141.000000 80.000000 36.000000 190.000000 36.600000
max 199.000000 122.000000 99.000000 846.000000 67.100000Note that the counts for our troublesome columns have changed as the (probably) missing fields are ignored - plus the means and standard deviations have changed.
Lets fill in the missing values with the average (mean) value for that feature.
And check again for missing values (there shouldn't be any):
Number of missing fields (post-fill)
------------------------------------
0 0
1 0
2 0
3 0
4 0
5 0
6 0
7 0
8 0
dtype: int64Now lets get the stats for the columns we filled-in:
Statistics (post-fill)
----------------------
1 2 3 4 5
count 768.000000 768.000000 768.000000 768.000000 768.000000
mean 121.681605 72.254807 26.606479 118.660163 32.450805
std 30.436016 12.115932 9.631241 93.080358 6.875374
min 44.000000 24.000000 7.000000 14.000000 18.200000
25% 99.750000 64.000000 20.536458 79.799479 27.500000
50% 117.000000 72.000000 23.000000 79.799479 32.000000
75% 140.250000 80.000000 32.000000 127.250000 36.600000
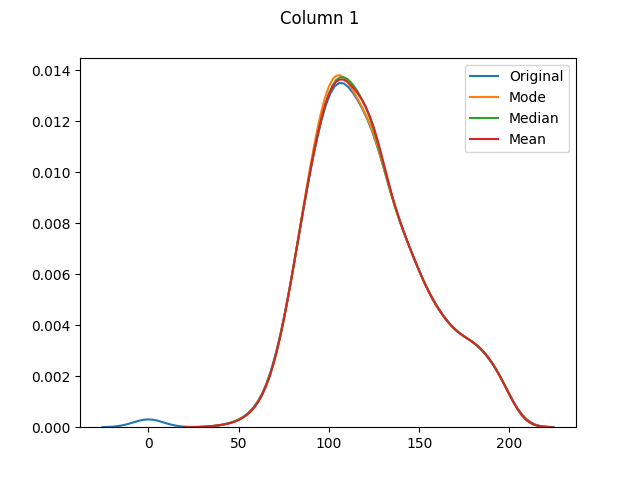
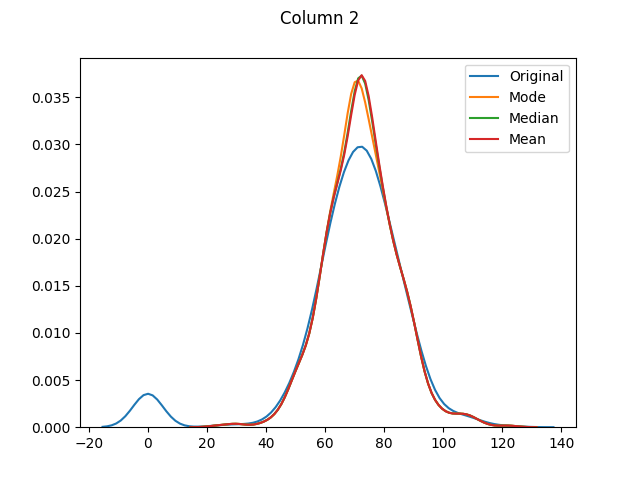
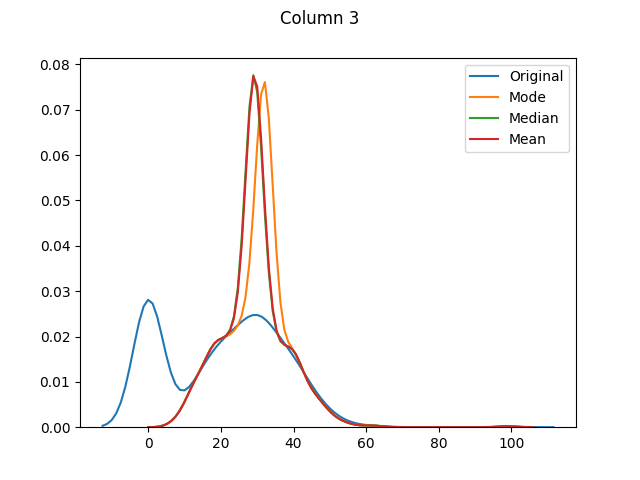
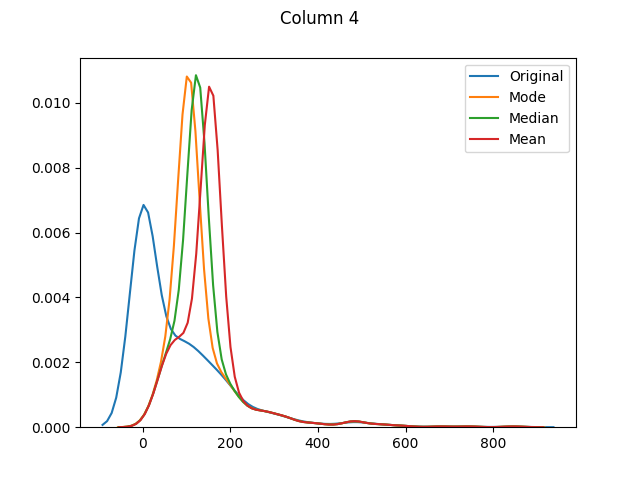
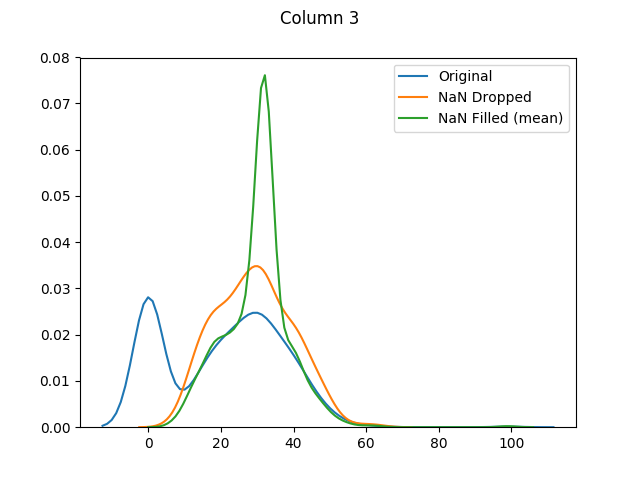
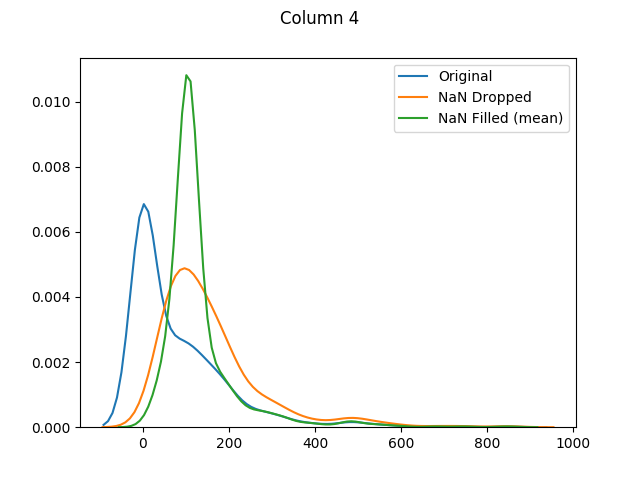
max 199.000000 122.000000 99.000000 846.000000 67.100000The means for columns 3 and 4 are different (in both of these columns zero was actually the mode - or most common value), but otherwise it's mainly the distributions that have shifted as the zero values have been adjusted:
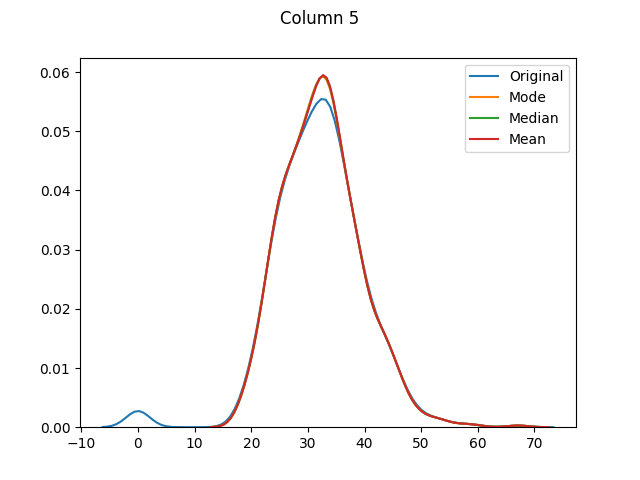
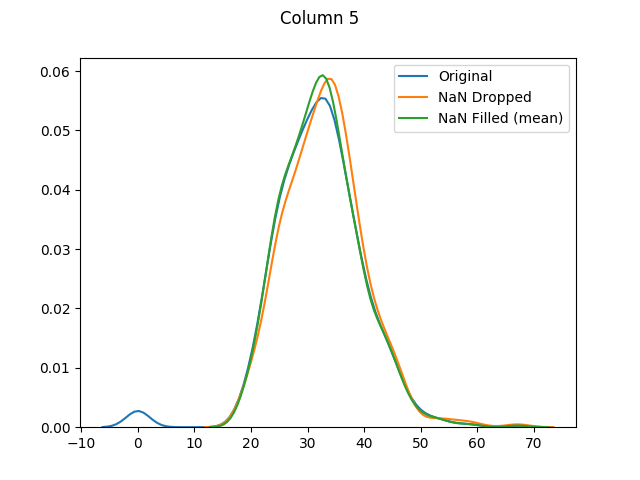
[Column 5 only had 11 missing values. As it is fairly normally-distributed, the mode, median and mean distributions seem to be almost identical.]
Note that we cannot use a dataset with NaN values for k-fold cross validation:
Accuracy (with NaN values)
--------------------------
/home/owner/.local/lib/python2.7/site-packages/sklearn/model_selection/_validation.py:542: FutureWarning: From version 0.22, errors during fit will result in a cross validation score of NaN by default. Use error_score='raise' if you want an exception raised or error_score=np.nan to adopt the behavior from version 0.22.
FutureWarning)
Input contains NaN, infinity or a value too large for dtype('float64').[Throws a ValueException, the value of which is shown.]
Now we will use the Pandas dropna function to drop any entries that contain NaN values.
Rows, columns (NaN values dropped) = (392, 9)
Statistics (NaN values dropped)
-------------------------------
1 2 3 4 5
count 392.000000 392.000000 392.000000 392.000000 392.000000
mean 122.627551 70.663265 29.145408 156.056122 33.086224
std 30.860781 12.496092 10.516424 118.841690 7.027659
min 56.000000 24.000000 7.000000 14.000000 18.200000
25% 99.000000 62.000000 21.000000 76.750000 28.400000
50% 119.000000 70.000000 29.000000 125.500000 33.200000
75% 143.000000 78.000000 37.000000 190.000000 37.100000
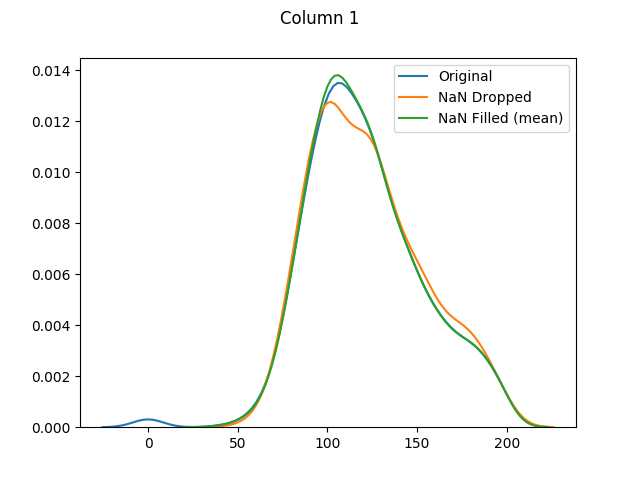
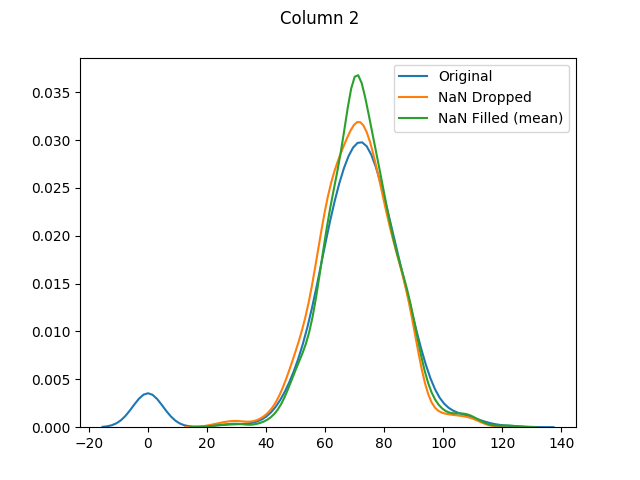
max 198.000000 110.000000 63.000000 846.000000 67.100000And almost half of our entries have now been dropped.
Let's compare our k-fold cross validation with dropped and filled values:
Accuracy (with NaN values dropped)
----------------------------------
0.78582892934
Accuracy (with NaN values filled)
---------------------------------
0.766927083333[These are exactly the same as the tutorial's published values.]
And finally let's use seaborn to graph our original values versus dropped values versus filled values:
Various useful links (and comments) are listed below.
https://scikit-learn.org/stable/modules/generated/sklearn.model_selection.cross_val_score.html
Will throw a ValueError for missing data:
ValueError: Input contains NaN, infinity or a value too large for dtype('float64').
http://seaborn.pydata.org/generated/seaborn.distplot.html
Will throw a ValueError for missing data:
ValueError: array must not contain infs or NaNs
http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.dropna.html
Default behavior is to drop entries where Any field is NaN.
http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.fillna.html
Fill in NA / NaN values.
http://pandas.pydata.org/pandas-docs/stable/generated/pandas.isnull.html
Detects missing values - such as NaN in numeric arrays, None or NaN in object arrays, NaT in datetimelike.
http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.mean.html
Note that the default value for skipna is True, which means invalid data will be ignored when calculating the column mean.
http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.median.html
Note that the default value for skipna is True, which means invalid data will be ignored when calculating the column median.
http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.mode.html
Note that multiple values may be returned for the selected axis. Also that the default value for numeric_only is False.
http://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.replace.html
Note that the value to be replaced can also be specified by a regex. Also that the default value for inplace is False.
missing data with pandas:
http://pandas.pydata.org/pandas-docs/stable/missing_data.html
missing data with sklearn:
http://scikit-learn.org/stable/modules/impute.html#impute
- Add a Snyk.io vulnerability scan badge
- Graph before and after (mean, median and mode) values
- Conform code to
pylint,pycodestyleandpydocstylestandards - Fix annoying
sklearnFutureWarning warnings - Generate a Monte Carlo style missing-data dataset and evaluate how it performs (in comparison to its non-missing-data original)
- Finish tutorial
I (mainly) followed this excellent tutorial:
http://machinelearningmastery.com/handle-missing-data-python/