chemmodlab contains a suite of methods for fitting machine learning models and for validating the resulting models:
-
ModelTrain()fits a series of classification or regression models to sets of descriptors and computes cross-validated measures of model performance. Repeated k-fold cross validation is performed with multiple, different fold assignments for the data ("splits"). -
MakeModelDefaults()makes a list containing the default parameters for all models implemented in ModelTrain so that they can be modified. -
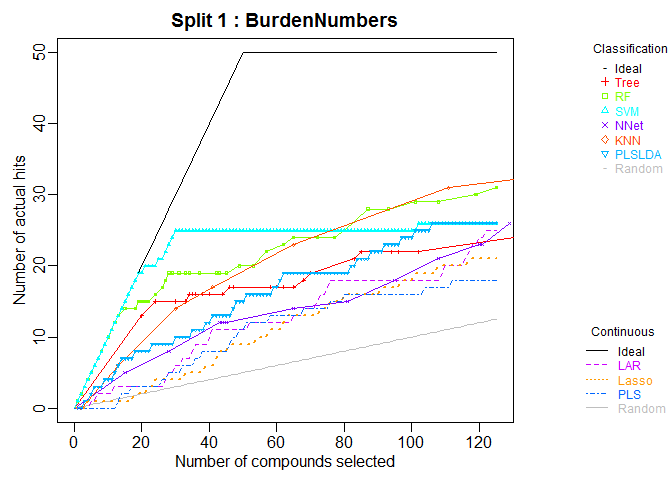
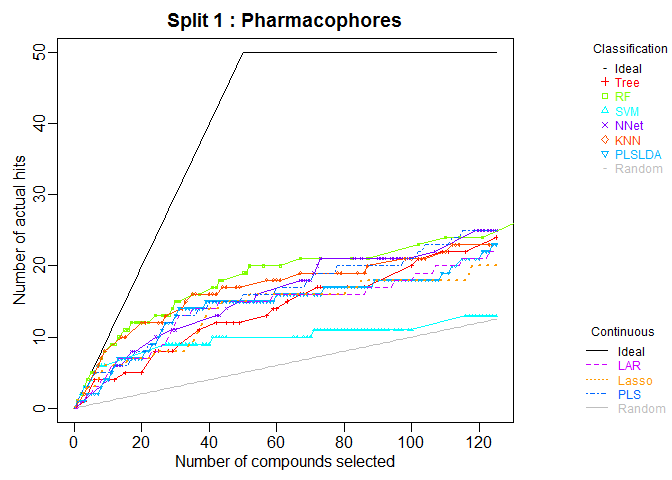
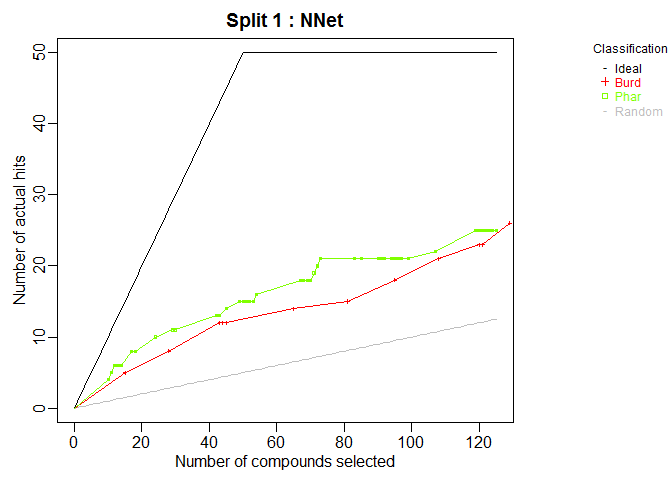
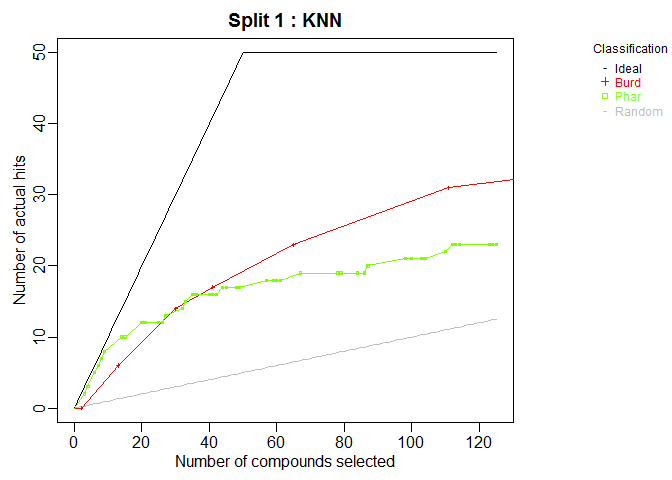
plot.chemmodlab()takes a chemmodlab object output by theModelTrainfunction and creates a series of accumulation curve plots for assesing model and descriptor set performance. -
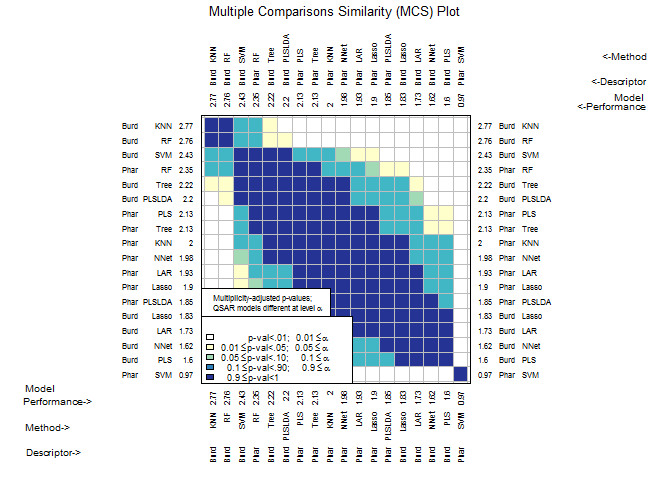
CombineSplits()evaluates a specified performance measure across all splits created byModelTrainand conducts statistical tests to determine the best performing descriptor set and model (D-M) combinations. -
Performance()can evaluate many performance measures across all splits created byModelTrain, then outputs a data frame for each D-M combination. -
chemmodlab()is the constructor for the chemmodlab object.The statistical methodologies comprise a comprehensive collection of approaches whose validity and utility have been accepted by experts in the Cheminformatics field. As promising new methodologies emerge from the statistical and data-mining communities, they will be incorporated into the laboratory. These methods are aimed at discovering quantitative structure-activity relationships (QSARs). However, the user can directly input their own choices of descriptors and responses, so the capability for comparing models is effectively unlimited.
# install from CRAN
install.packages("chemmodlab")
# Or use the development from GitHub:
# install.packages("devtools")
devtools::install_github("jrash/chemmodlab")library(chemmodlab)
data(aid364)
cml <- ModelTrain(aid364, ids = TRUE, xcol.lengths = c(24, 147),
des.names = c("BurdenNumbers", "Pharmacophores"))
plot(cml, splits = 1, meths = c("NNet", "KNN"))CombineSplits(cml, metric = "enhancement", m = 100)