Human Taenia solium force-of-infection modelling project
Project to fit catalytic models to human Taenia solium age-(sero)prevalence data to estimate per-capita seroconversion / infection rates (force-of infection; FoI) and seroreversion / infection loss rates.
Table of contents
Background
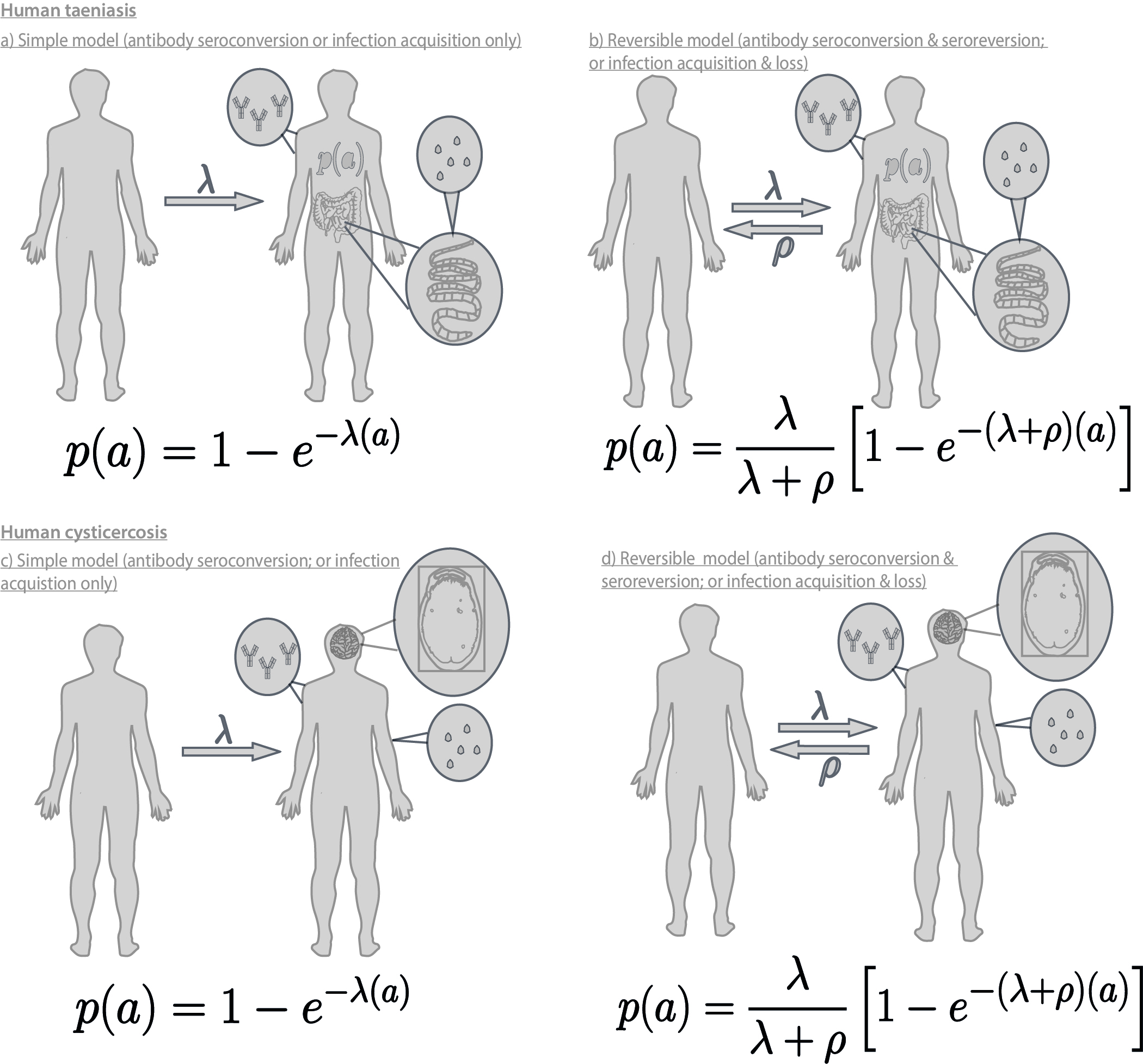
The FoI describes the average (per capita) rate at which susceptible individuals seroconvert (become Ab-positive) or become infected. Catalytic models, first proposed for fitting to epidemiological data by Muench (1934), can be fitted to age-(sero)prevalence curves to estimate the FoI. A reversible configuration of the model can also be used to estimate the (per-capita) rate at which individual serorevert (become Ab-negative) or clear infection.
This project presents the first analysis to estimate these rates for T. solium human cysticercosis and human taeniasis, across a range of epidmeiological settings, after identifying suitable age-(sero)prevalence data from a systematic literature review. Cataltyic models are fitted using a bayesian framework, incorporating uncertainty in diagnostic performance (sensitivity and specificty of each test used) using the formula given by Diggle, (2011). Markov chain Monte Carlo (MCMC) simulations are used to estimate posterior distributions for parameters of interest (FoI, seroreversion/infection loss and diagnostic sensitvity and specificty). Our analysis therefore assesses global variation in these important epidemiological quantities to inform development of transmission models, and to support control efforts.
We provide code to support this analysis wtihin this repository.
Cataltyic models used to estimate rates in the paper are detailed in the following figure (full colour figure & parameter tables will become available after publication):
References:
Muench H (1943). Derivation of Rates from Summation Data by the Catalytic Curve. Journal of the American Statistics Association 29:25–38. doi: 10.2307/2278457. https://www.jstor.org/stable/2278457
Diggle P (2011). Estimating Prevalence Using an Imperfect Test. Epidemiology Research International. doi: 10.1155/2011/608719. https://www.hindawi.com/journals/eri/2011/608719/
Code guide
The master script is split into:
- fitting to single datasets
- fitting to multiple datasets (this will estimate FoI/seroreversion or infection loss from each dataset, with a joint sensitivity and specifcity across datasets)
Data availability
All age-(sero)prevalence data are available in the following data repository: http://doi.org/10.14469/hpc/10047 at Imperial College London. Original data for two datasets available (under the Creative Commons Attribution License; CC BY 4.0) from the International Livestock Research Institute open-access repository (http://data.ilri.org/portal/dataset/ecozd) referenced in Holt et al. (2016) and University of Liverpool open-access repository (http://dx.doi.org/10.17638/datacat.liverpool.ac.uk/352) referenced in Fèvre et al. (2017).
Other related work
This is a complementary study to the pig cysticercosis FoI analysis conducted by this group and published in Scientific Reports:
Together, these studies can be used to parameterise both the pig and human elements of a T. solium transmission model (where FoI estimates for pig and human infection exist for similar settings).