AMBIA (Accurate Mouse Brain Image Analysis) is a tool to fascilitate the analysis of 2D histological Mouse brain images. It is written purely in python and has a modular design and consists of the following modules:
- Cell detection Module
- Localization Module
- Atlas generator module
- Registration Module
AMBIA is intentionally designed with few requirements. You can easily run AMBIA on your local computer. Ofcourse computers with higher ram and stronger CPUs make AMBIA run faster. Following a the requirements before you install AMBIA:
- Python > 3.7
-
Install python 3.9
https://www.python.org/downloads/release/python-3910/
Download and run the Windows installer (64-bit)
In your installation check Add to PATH -
Install Git https://git-scm.com/download/win
Download and install the 64-bit Git for Windows Setup -
Open Command prompt go the the path that you want to install AMBIA
cd the_path_to_install_ambia
4- Run the command below to clone AMBIA.
Git clone https://github.com/mrymsadeghi/AMBIA.git -
Use the requirements.txt file provided in the package to install the necessary python packages with optimal versions.
cd AMBIA
pip install -r requirements.txt -
Run AMBIA
cd mb_gui\src
python Main.py
You can create a desktop shortcut icon:
- Open a notebook. Write the 3 lines below
C:
cd the_path_for_ambia\AMBIA\mb_gui\src
Python Main.py - Save it with any name but the the suffix .bat not .txt. And save this file anywhen (e.g. on the desktop) as the AMBIA icon. Replace C in the codes below by the drive letter AMBIA is installed in.
The files and folders in the AMBIA Git repository consists of the following folers:
- Gui Atlases
- Ambia_core
- models
- Processed
Keep this folder structure in your system for AMBIA to work correctly.
Here is an explanation of the content and functionality of each folder:
This folder contains the color coded coronal atlases of adult mouse brain or P56 mouse brain. In the file Static_Switches.py if you choose atlas_type = Adult, AMBIA will use Adult atlases in this folder. Also it is noteworthy that the color codes of each atlas is contained in the folder Ambia_core/src/atlas_codes.
This folder contains all the python code for the Cell detection, Localization, Atlas generator and Registration Module, as well as the GUI. The file Ambia_core/src/Switches_Static.py contatins parameters that are editable by the users. Parameters such as for choosing the atlas type (atlas_type), turning auto registration on or off (auto_registration) and etc.
This folder contains three different type of data
- Deep learning trained models for the localization module. Including the SL predictor, QL predictor, Group A segmentation modes
- Excel sheet templates in which Ambia exports data
- Files required for generating tilted 2D allen mouse brain atlas from the 3D atlas in the Allen_files folder
This is where the files and reports of the analysis of your mouse brain slices will be saved. Also as an example, we provide analysis of two mouse brain slices from two different slides are in this folder.
If you use our software for your research, we kindly request that you cite our corresponding paper in any resulting publications. The reference for the paper is:
Sadeghi, Maryam, et al. "Localization and Registration of 2D Histological Mouse Brain Images in 3D Atlas Space." Neuroinformatics (2023): 1-16.
Citing this paper helps us to continue developing and maintaining this project. It also ensures that the scientific community recognizes the effort we have put into creating this software. It's important to give credit where it's due, and we appreciate your support in acknowledging our work.
Here are some screenshots of AMBIA GUI showing different stages of analysis with a mrxs format slide image.
Loading the Slide image
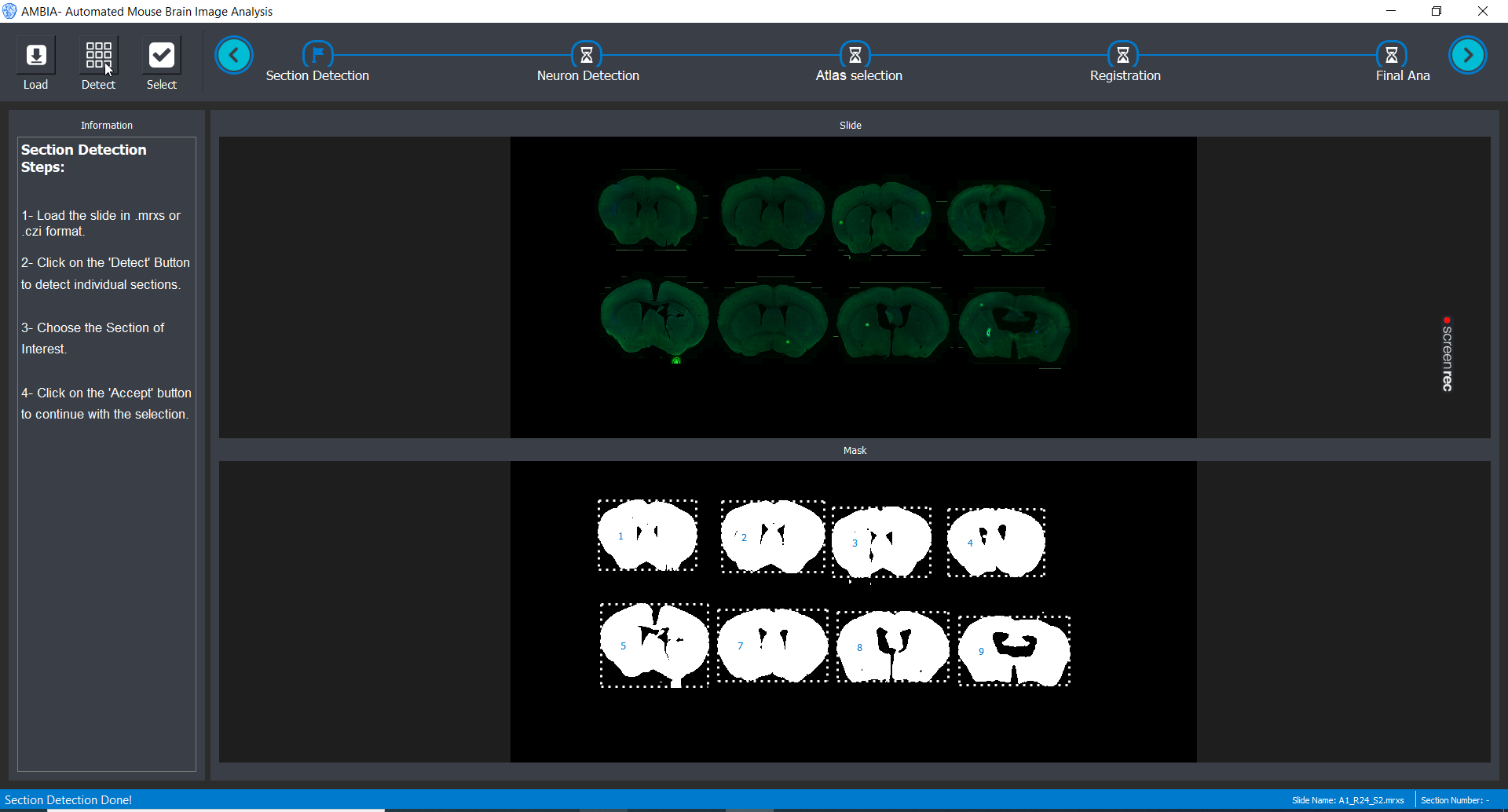
Slide image is loaded. AMBIA detects different mouse brain slices, and now the user can choose which slice to analyse.
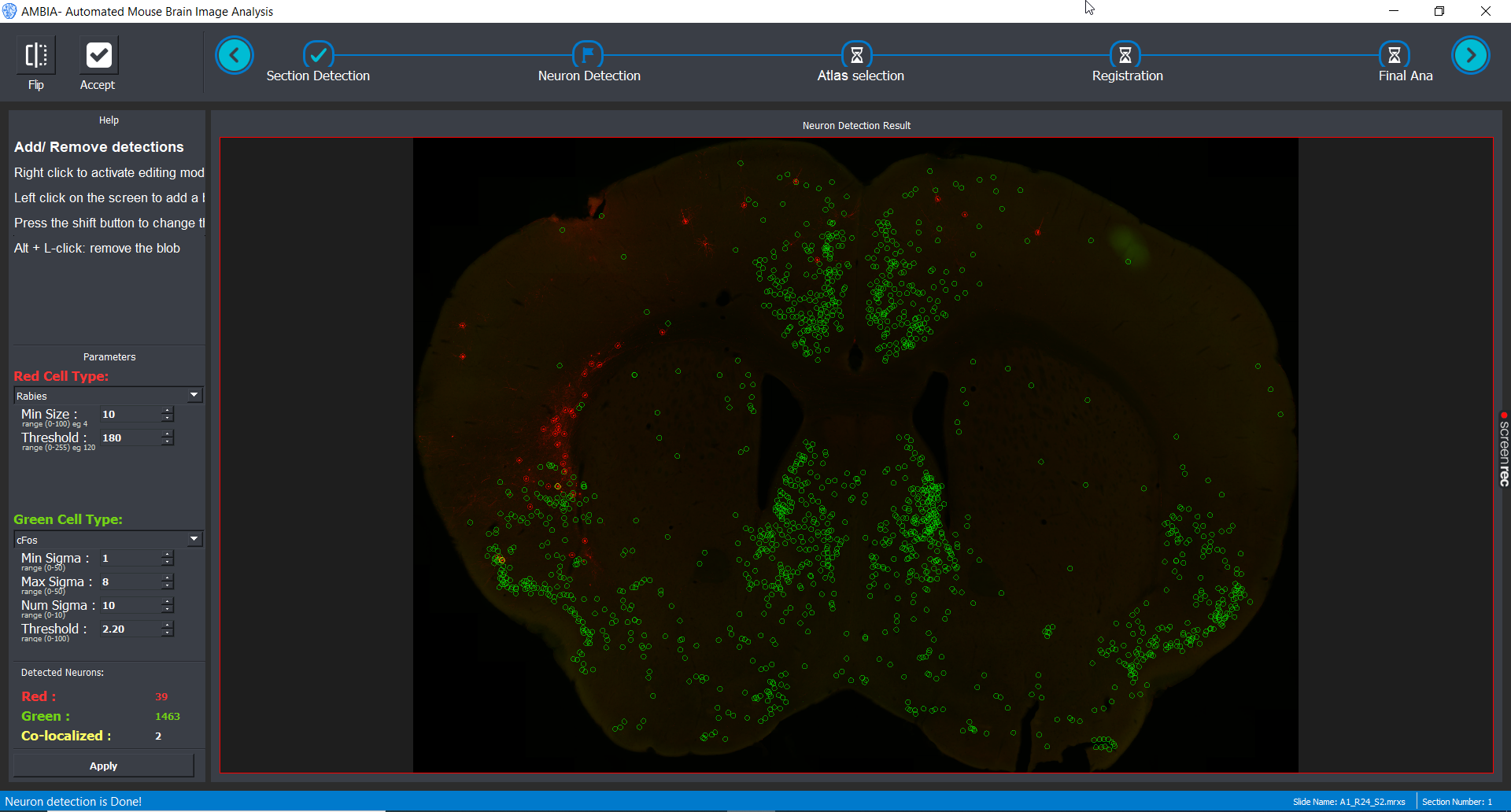
Cell Detection
Different methods can be implemented in the cell detection module. Here as default two methods for each channel is provided. The user can choose between methods for each of the green and red channels and parameters can be adjusted in the GUI
Localization
The selected slice can now be registered to a mouse brain atlas. The user can either choose a standard coronal allen atlas based on its number. Or by toggling the "Tilted atlas" switch in the GUI, you can use the deep learning localization module to detect the position of the slice and generate a custom atlas.
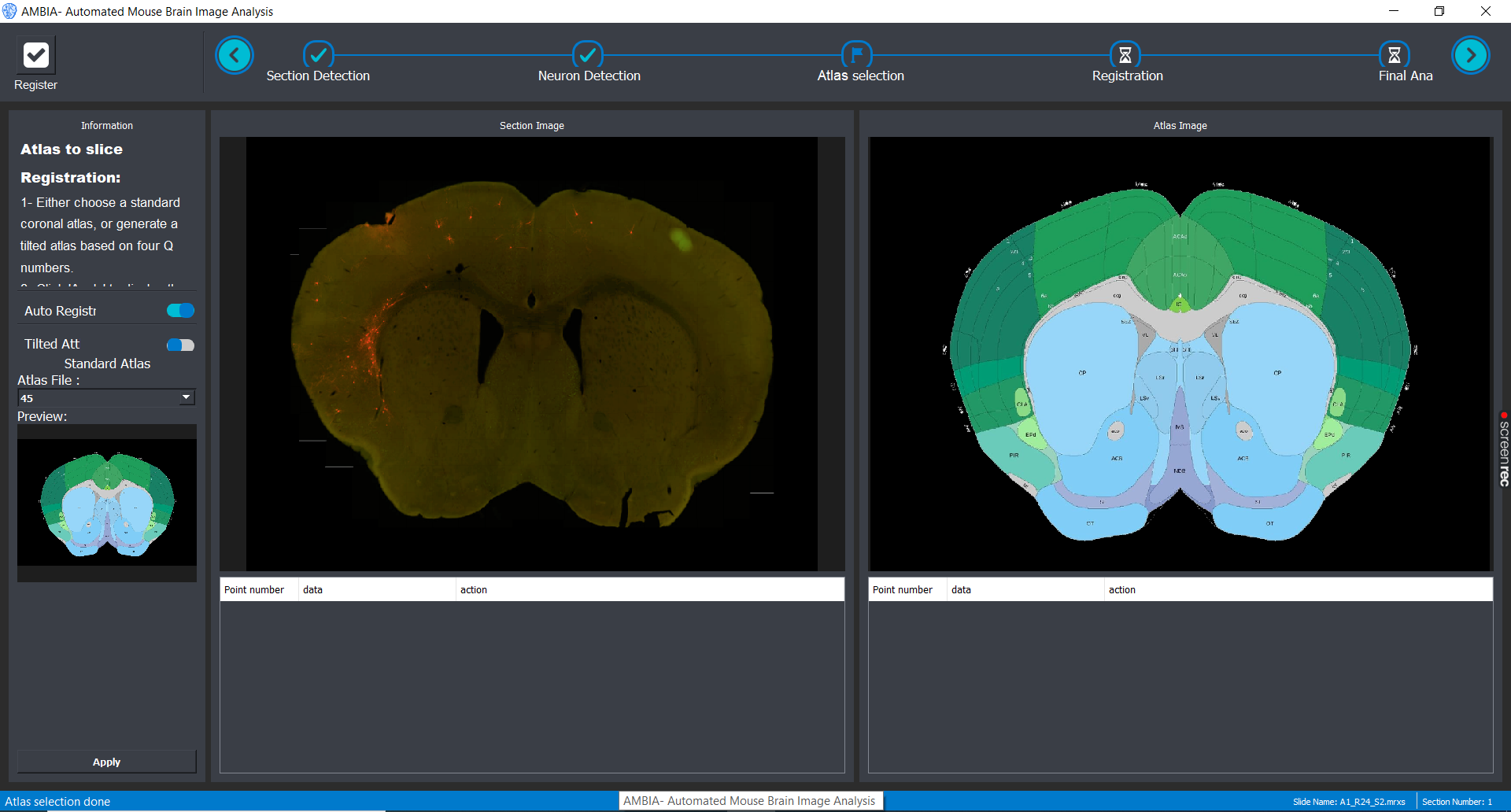
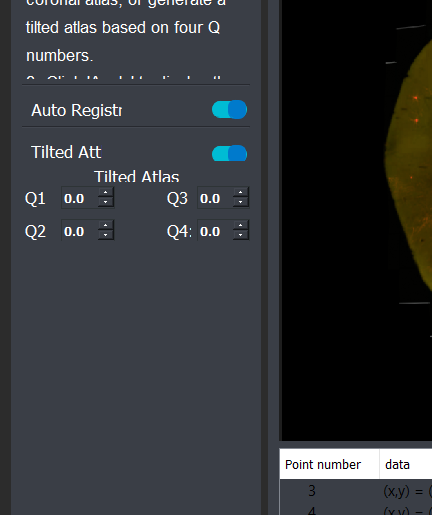
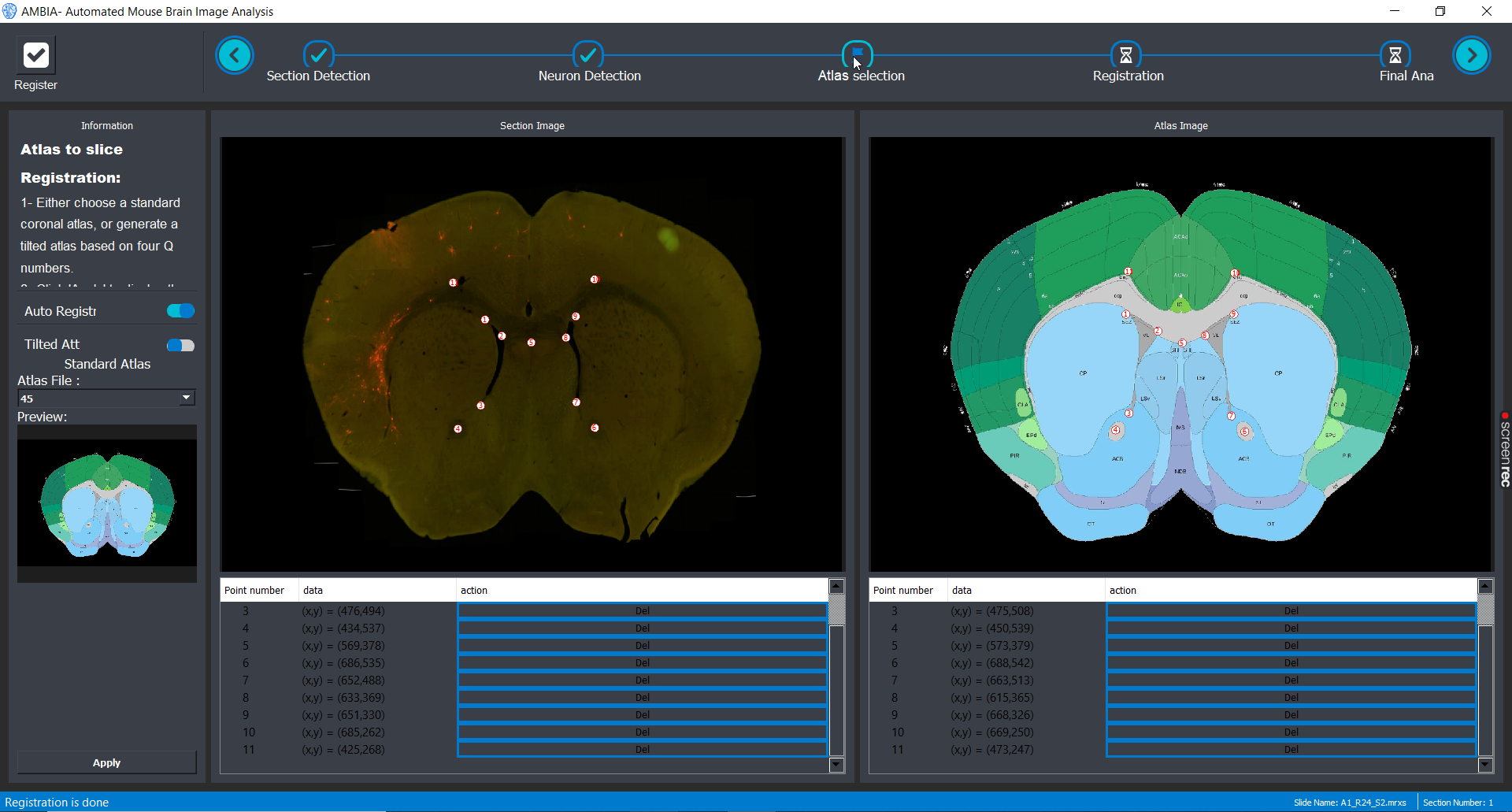
Registration
After choosing or generating an atlas, you can toggle auto registration switch on to perform automatic ardent registration. And later choose landmarks to refine the registration.
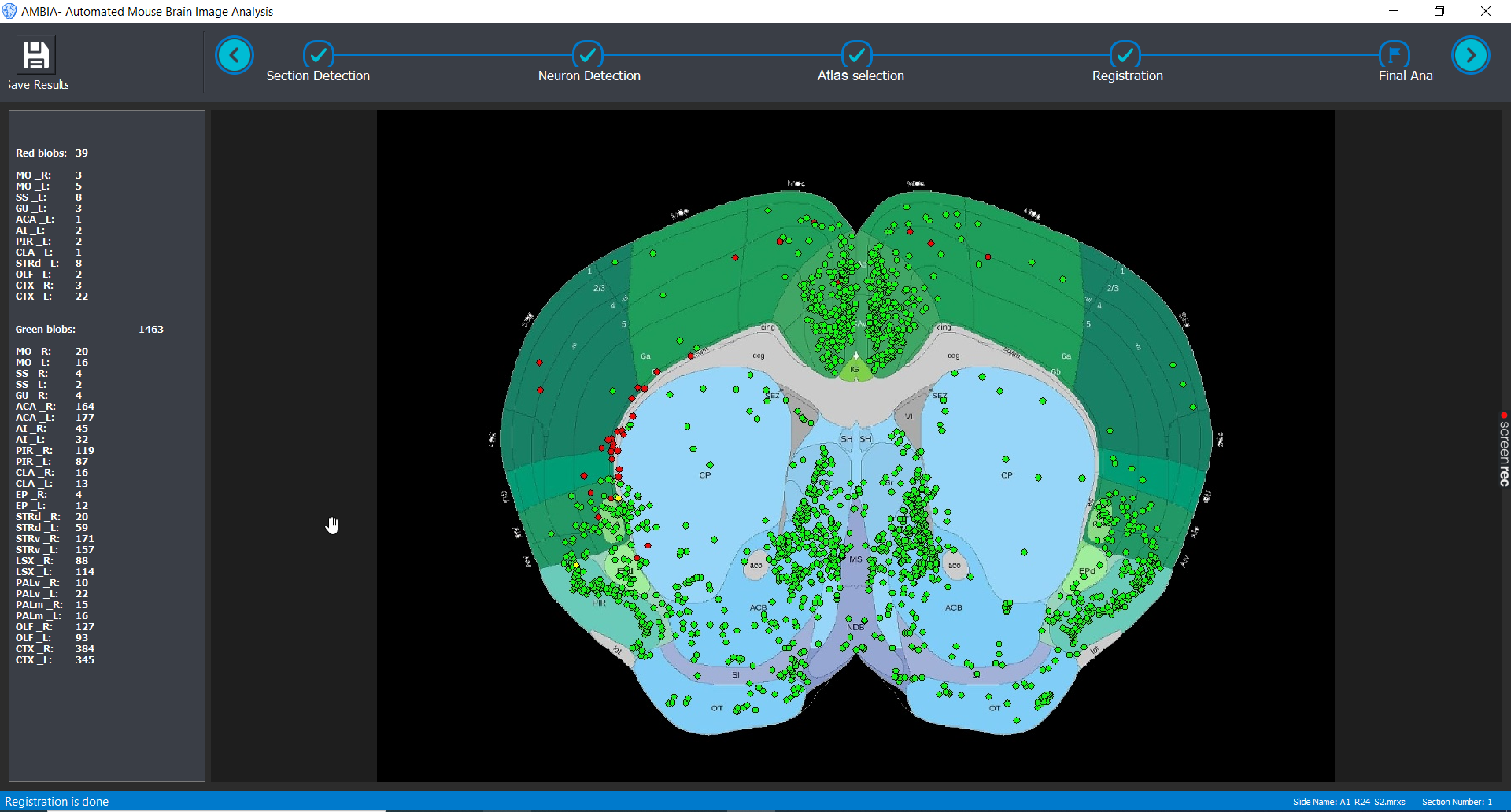
Analysis Report
AMBIA demostrates visually where the detected cells are located in the registered mouse brain slice on the annotated atlas. Also a report is generated in the form of an Excel sheet and a text file which reports the number of detected cells in each brain region.
You can find video tutorials on how to install and use AMBIA in the following links:
Installation and how to start (Available soon)
How to Use AMBIA (Available soon)
How to modify the code of AMBIA (Available soon)
-
Sadeghi, Maryam, et al. "Localization and Registration of 2D Histological Mouse Brain Images in 3D Atlas Space." Neuroinformatics (2023): 1-16. https://link.springer.com/article/10.1007/s12021-023-09632-8
-
Sadeghi, Maryam et al. "Automatic 2D to 3D localization of histological mouse brain sections in the reference atlas using deep learning". In Medical Imaging 2022: Image Processing (Vol. 12032, pp. 718-724). SPIE https://doi.org/10.1117/12.2604231
Development of AMBIA on Gitlab since 2020 here
Ardent package Github here