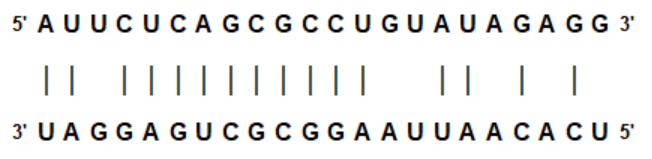A home for R functions I use myself to visualize genomic features. Will grow over time. Should serve as a starting point for own representations and compilations (multi panel plots).
Each function should do three things at its core:
- easy to use (data.frame or string/vector as input - no fancy objects)
- lightweight
- no dependencies - plain base R
Single functions can be sourced directly using:
# 'geneset.r' example
library(devtools)
source_url("https://github.com/mschemmel/genfuncR/blob/main/geneset.r?raw=TRUE")library(grid)
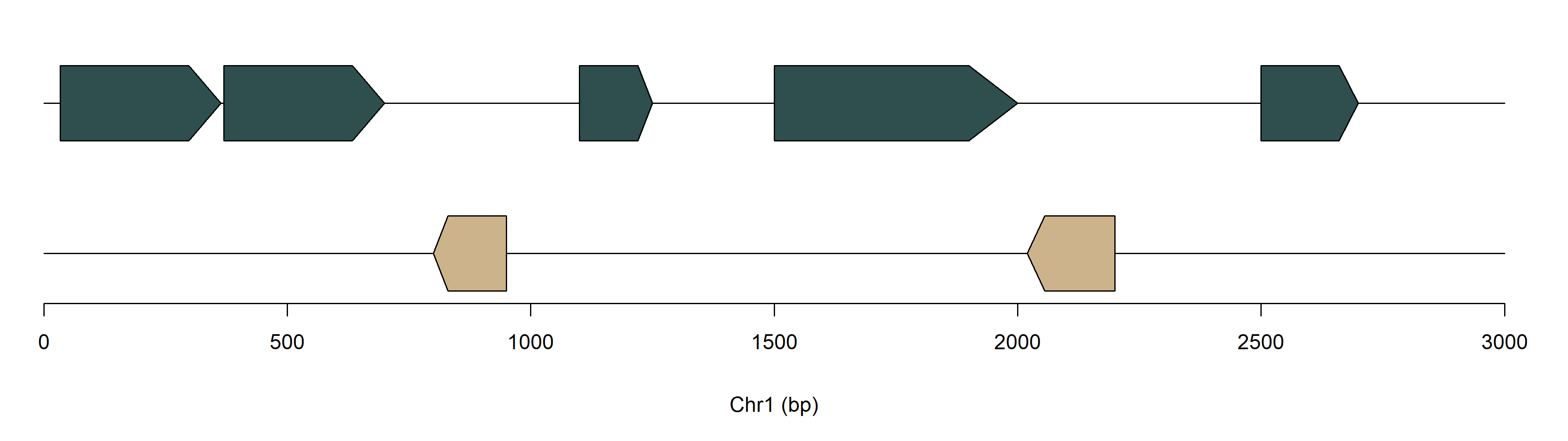
genes <- data.frame(chr = c("Chr1", "Chr1", "Chr1", "Chr1", "Chr1", "Chr1", "Chr1"),
start = c(34, 370, 800, 1100, 1500, 2020, 2500),
end = c(364, 700, 950, 1250, 2000, 2200, 2700),
strand = c("+", "+", "-", "+", "+", "-", "+"))
# basic example
geneset(geneTrack(genes))Basic example
# more advanced example...coming soon againlibrary(grid)
# basic example
nucleotides <- paste0(sample(c("A","T","G","C"), 1700, replace = TRUE), collapse = "")
dna2img(nucleotides)Basic example
library(grid)
mirna <- "AUUCUCAGCGCCUGUAUAGAGG"
target <- "UAGGAGUCGCGGAAUUAACACU"
# basic example
mirnali(mirna, target)Basic example
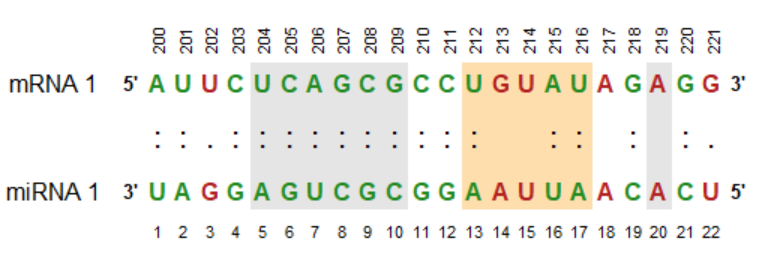
# more advanced example
mirnali(mirna,
target,
mirna_name = "miRNA 1",
target_name = "mRNA 1",
alignment_type = ":",
match_color = "forestgreen",
mismatch_color = "firebrick",
highlight_area = list("A" = c(5, 10), "B" = c(13, 17), "C" = c(20, 20)),
highlight_color = list("A" = "gray90", "B" = "navajowhite", "C" = "gray90"),
target_position_label = c(200:(199 + nchar(target))),
target_position_label_rot = 90)More advanced example