Still Under developing..
It's quite attractive to use amount of data including TCM(中医药, Tradition Chinese Medicine), formulas(药方), ingredients, targets, and related disease to find active natural compounds or new drugs from our mother land. However, as a beginner of bioinfomatic, I found most of Tradition Medicine databases (such as herbs, TCMSP, ETCM) are not very friendly to us. They don't service a interface for us to download all of their data. Or only give a single dataframe without same columns to combine each other.
So, I am going to use R as my sword for taking this big challenge. Also, I can practice my web crawler skills.
In this project, I just want to crawl some main TCM database. Correct some mistakes, clean wrong data, reformat them into concise and elegant tibble/data.frame format.
You can see the 01-preparation.R file in the main directory. Besides, you need do more. Here, we use Selenium Server in java environment.
The software we need are: java, Selenium Server, chromedriver, Google Chrome, R and R Studio

You can install the latest and free one in https://www.java.com/
-
For Windows: open your cmd, enter "java -version"
IF you don't have java in your path, you should add them. (in case of you don't have java, please install it)
-
For Windows: answer in: https://explainjava.com/set-java-path-and-java-home-windows/
-
For Mac os: https://explainjava.com/set-java-home-mac-os
Here I quote a passage from RSelenium cran docs:
Selenium Server is a standalone java program which allows you to run HTML test suites in a range of different browsers, plus extra options like reporting.
Our R packages run basically by this server.
You can get it from: http://www.seleniumhq.org/download/ Or http://selenium-release.storage.googleapis.com/index.html
It's a driver we need for Chrome in order to run Selenium server. http://chromedriver.storage.googleapis.com/ for mainland user, here is mirror (https://npm.taobao.org/mirrors/chromedriver)
Notably, we should download correct version of driver.
You can check it in:
After downloading, you should run both Selenium Server and Chromedriver in your terminal or cmd:
java -Dwebdriver.chrome.driver="/Path/to/your/chromedriver" -jar /Path/to/your/selenium-server-standalone-3.141.59.jar
At the start, I mentioned 01-preparation.R.
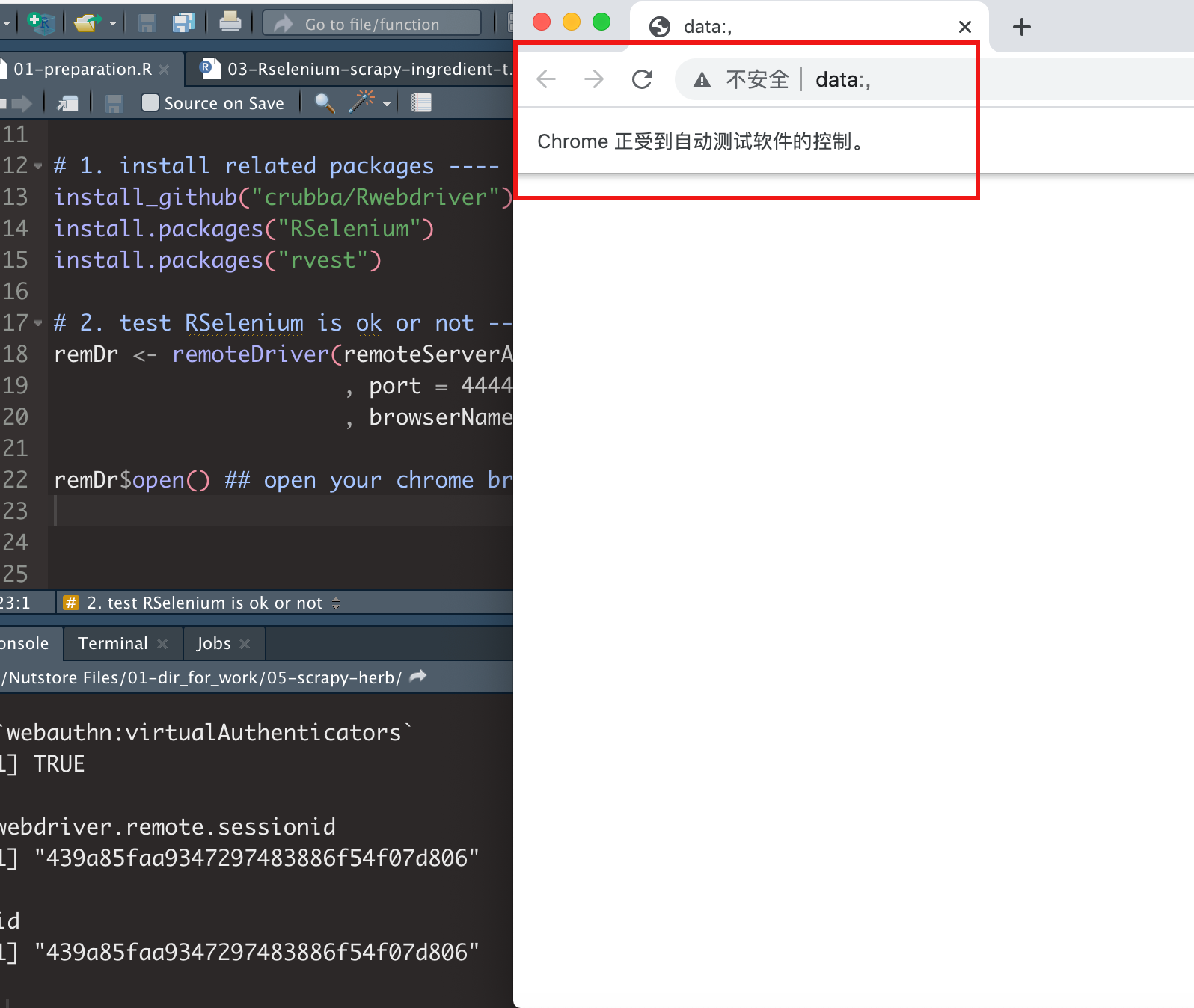
If you successfully run RSelenium in R, you will see a browser opened.
Have a try and enjoy it!
It's finished by RSelenium remote control!
Here is a comprehensive tutorial for Rselenium: https://docs.ropensci.org/RSelenium/articles/basics.html
Chinese Version By me: https://www.yuque.com/mugpeng/rr/hg7gu8
Each step has its own R script.
If you successfully run 01-preparation.R, it will be easy for you.
I split my project code into two part:
- main script, each represent a specific work.
- Rscript for sourcing, which means they are mainly pre-built functions and only directly used from main script. Including related-function(small function) and main-function(big one).
If there is no bug, the happy result is you only need to run the main script and just wait for your output.
All crawl output will be stored in a list:
> my_list[[172]]
$my_ID
[1] "HBTAR000172"
$my_url
[1] "http://herb.ac.cn/Detail/?v=HBTAR000172&label=Target"
$my_page_ID
[1] "HBTAR000172"
$my_child_table
# A tibble: 2 × 3
`Ingredient id` `Ingredient name` `Database sources`
<chr> <chr> <chr>
1 HBIN018385 BGC SymMap:SMIT07197; TCMSP:MOL00…
2 HBIN045073 sucrose SymMap:SMIT00425; TCMID:20430…
$my_status
[1] "OK"
$my_type
[1] "Ingredient-Target"
each element represent:
- my_ID && my_page_ID : ID we designed for crawling and ID from web recording.
- my_url: Url for this record.
- my_child_table: tibble of related data.
- my_status: Check if row of our table is same with row recorded in the web page, and others. IF shows ok, this record is seems to be correctly crawled.
- my_type: Ingredient-Target, or Ingredient-Disease.
- my_source: herb, ETCM, etc.
$ tree -L 1 backup/; tree -L 1 selenium_file/
backup/
├── disease
├── herbs
└── targets
3 directories, 0 files
selenium_file/
├── LATEST_RELEASE_92
├── chromedriver
├── chromedriver.exe
└── selenium-server-standalone-3.141.59.jar
In backup, there are my temporal Rdata file by my web crawler. I will combine each of them into a single data.frame soon. And you can use file in selenium_file for setting ur Selenium Server and Chromedriver, and my Chrome version is 92.0.
-
herb(本草图鉴): http://herb.ac.cn/ A high-throughput experiment- and reference-guided database of traditional Chinese medicine.
-
ETCM(The Encyclopedia of Traditional Chinese Medicine): http://www.tcmip.cn/ETCM/index.php/Home ETCM includes comprehensive and standardized information for the commonly used herbs and formulas of TCM, as well as their ingredients. To facilitate functional and mechanistic studies of TCM, ETCM provides predicted target genes of TCM ingredients, herbs, and formulas. A systematic analysis function is also developed in ETCM, which allows users to explore the relationships or build networks among TCM herbs, formulas, ingredients, gene targets, and related pathways or diseases. ETCM is free for academic use and the data can be conveniently exported.
-
TCMSP(Traditional Chinese Medicine Database and Analysis Platform): https://tcmsp-e.com/ TCMSP is a unique systems pharmacology platform of Chinese herbal medicines that captures the relationships between drugs, targets and diseases.
-
TCMID(Traditional Chinese Medicine Information Database):http://bidd.group/TCMID/ Linking TCM Data to Human Genomic Profiles
After crawling herbs, I will re-crawl the wrong data which doesn't recorded as "OK". Besides, I will clean the data and reformat it into a new tibble/data.frame format data with my special barcode. If it possible, I will crawl more database to enhance the reliability and content of my package/database.
Furthermore, I will develop a R package for storing, sharing, achieving, visualizing TCM database data.