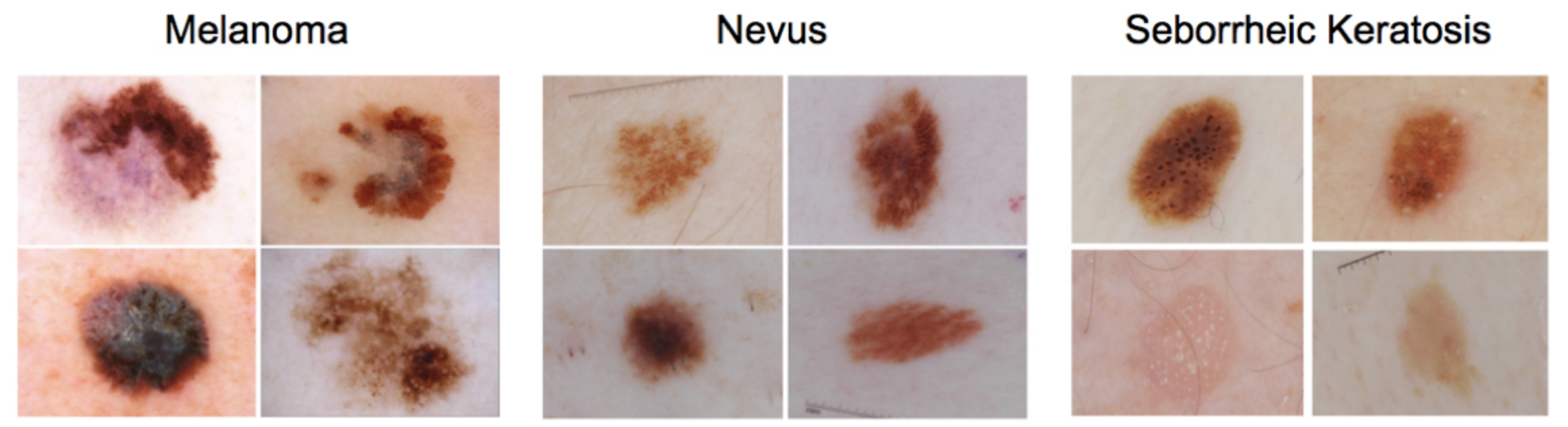
This repository contains the definition and evaluation of a Convolutional Neural Network (CNN) which aims to classify skin lesion images into three categories:
- Melanoma: malign cancer, one of the deadliest.
- Nevus: benign skin lesion (mole or birthmark).
- Seborrheic keratosis: benign skin tumor.
Additionally, the images are compressed with an Autoencoder and visualized using T-SNE.
The motivation of the project comes from two sources that caught my attention:
- The Nature paper by Esteva et al., in which the authors show how a CNN architecture based on the Inception-V3 network achieves a dermatologist-level classification of skin cancer.
- The 2017 ISIC Challenge on Skin Lesion Analysis Towards Melanoma Detection.
I decided to try one task of the 2017 ISIC Challenge; to that end, I forked the Github repository udacity/dermatologist-ai, which provides som evaluation starter code as well as some hints on the challenge.
Overview of contents:
Although the 2017 ISIC Challenge on Skin Lesion Analysis Towards Melanoma Detection is already closed, information on the challenge can be obtained from the official website.
The challenge had three parts or tasks:
- Part 1: Lesion Segmentation Task.
- Part 2: Dermoscopic Feature Classification Task.
- Part 3: Disease Classification Task.
The present project deals only with the third part or task, which is evaluated with the ROC-AUC of three cases:
- The binary classification between benign (nevus and seborrheic keratosis) vs. malign (melanoma),
- The binary classification between skin lesions of origin in the melanocyte skin cells (nevus and melanoma) vs keratinocyte skin cells (seborrheic keratosis).
- The mean of the two above.
I have created a model that performs a multi-class classification and computed from its inferences the metrics of the 3 cases, but I have mainly focused on the first case: benign vs. malign. Additionally, I have defined an Autoencoder which generates compressed representations of the images; with them, I have applied a T-SNE visualization to the test split.
The challenge organizers published an interesting summary of the results and insights from the best contributions in this article; some interesting points associated with the third part/task are:
- Top submissions used ensembles.
- Additional data were used to train.
- The classification of seborrheic keratosis seems the easiest task.
- Simpler method led to better performance.
Some of the works that obtained the best results are:
- Matsunaga K, Hamada A, Minagawa A, Koga H. “Image Classification of Melanoma, Nevus and Seborrheic Keratosis by Deep Neural Network Ensemble”.
- Díaz IG. “Incorporating the Knowledge of Dermatologists to Convolutional Neural Networks for the Diagnosis of Skin Lesions”. Code.
- Menegola A, Tavares J, Fornaciali M, Li LT, Avila S, Valle E. “RECOD Titans at ISIC Challenge 2017”. Code.
I downloaded the dataset from the links provided by Udacity to the non-committed folder data/, which is subdivided in the train, validation and test subfolders as well as class-name subfolders:
- training data (5.3 GB)
- validation data (824.5 MB)
- test data (5.1 GB)
The images originate from the ISIC Archive.
The project folder contains the following files:
skin_lesion_classification.ipynb # Project notebook 1: Classification
dataset_structure_visualization.ipynb # Project notebook 2: T-SNE Visualization
README.md # Current file
Instructions.md # Original project instructions from Udacity
data/ # Dataset
images/ # Auxiliary images
requirements.txt # Dependencies
results.csv # My results with the model from notebook 1
LICENSE.txt # License
models/ # (Uncommitted) Models folder
get_results.py # (Unused) Script for generating a ROC plot + confusion matrix
ground_truth.csv # (Unused) True labels wrt. 3 cases/tasks of part 3
sample_predictions.csv # (Unused) Example output wrt. 3 cases/tasks of part 3
The most important files are the notebooks, which contain the complete project development. The dataset is contained in data/, but images are not committed.
Note that there are some unused files that come from the forked repository; the original Instructions.md explain their whereabouts.
Install the dependencies and open the notebooks, which can be run independently and from start to end:
skin_lesion_classification.ipynb: CNN model definition, training and evaluation with frozen ResNet50 as backbone.dataset_structure_visualization.ipynb: Convolutional Autoencoder definition and training; the encoder is used to generate compressed image representations, which are used to visualize the dataset applying T-SNE.
Note that if you run the notebooks from start to end you'll start training the models at some point, which might take several hours (each model took 6-8 hours on my Macbook Pro 2021 M1 with the current settings).
The project has a strong research character; the code is not production ready yet 😉
A short summary of commands required to have all in place with conda:
conda create -n derma python=3.6
conda activate derma
conda install pytorch torchvision -c pytorch
conda install pip
pip install -r requirements.txtBoth models produce very bad results; however, the research framework is now built to explore more appropriate approaches, as outlined in the improvements section 😉
- Current versionI trained this version on Google Colab Pro with a Tesla T4 for 4-5 hours (50 epochs)
- This version is better than the previous one: Frozen ResNet50 trained for 20 epochs
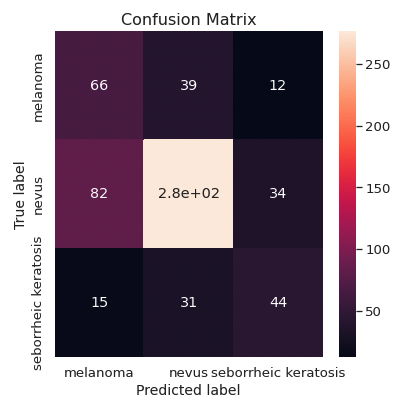
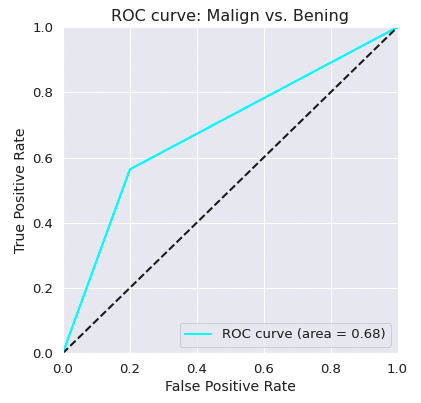
- The classifier has a ROC-AUC of 0.68 for the simplified classification benign vs. malign.
- Melanoma and seborrheic keratosis are often predicted as other skin lesions.
- The recall for the malign vs. benign case is 0.58.
- The autoencoder is not able to meaningfully compress the images, and hence, the encoded image representations are not
- The autoencoder is biased: it underfits the dataset, because the learning curves decrease fast at the begining and don't change much later on.
- The transformed images are quite different/low resolution compared to the original ones; it's difficult for the compressed representations to contain enough information to distinguish between classes.
- The transpose convolutions create checkerboard artifacts, as reported by Odena et al..
- As expected, the T-SNE transformation in 2D doesn't shown clearly differentiated point clusters.
|
Input to Autoencoder 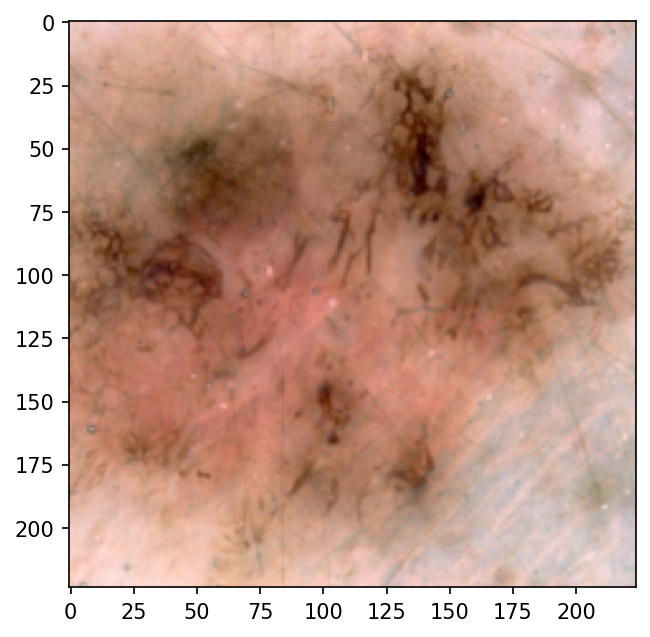
|
Output from Autoencoder 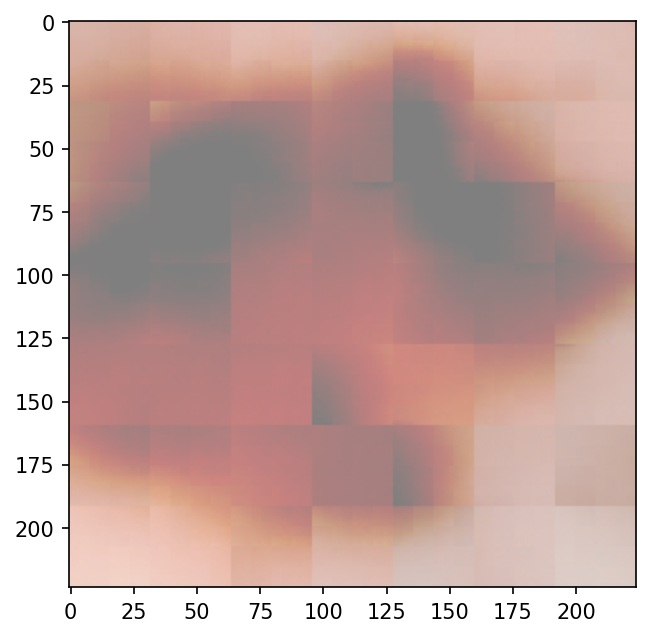
|
- Similarly as Esteva et al. did, I should fine tune the backbone, i.e., the weights of the backbone should be optimized for the dataset, too.
- Train it longer on Google Colab Pro or on a device with a more powerful GPU.
- Try with a learning rate scheduler, using a larger learning rate at the beginning
- Remove one fully connected layer in the classifier and check what happens
- Probably a strong data augmentation is needed; I can consider taking more images from the ISIC database
- Add more depth to the filters.
- Add linear layers in the bottleneck.
- Increase the number of parameters one order of magnitude and see if the behavior improves (i.e., 7M parameters).
- Use upsampling instead of transpose convolution (to solve the checkerboard artifacts). Example.
- It probably makes more sense to use the train split for the T-SNE visualization; however, I would need to set shuffle=False in the data loader to be able to track sample filenames easily.
- If the T-SNE visualization shows distinct clusters, I can try to find similar images simply by using the dot product distance between image vectors.
This repository was forked from udacity/dermatologist-ai and modified to the present status following the original license from Udacity. Note that the substantial content doesn't come from Udacity.
Mikel Sagardia, 2022.
No guarantees.