Clinical/Genetic Condition-based Model for Generating Brain T1 MRI Changes According to Alzheimer's Disease Progression
- Alzheimer's disease is a leading cause of dementia and the lack of effective treatment options highlights the critical need for early diagnosis and prediction of disease progression.
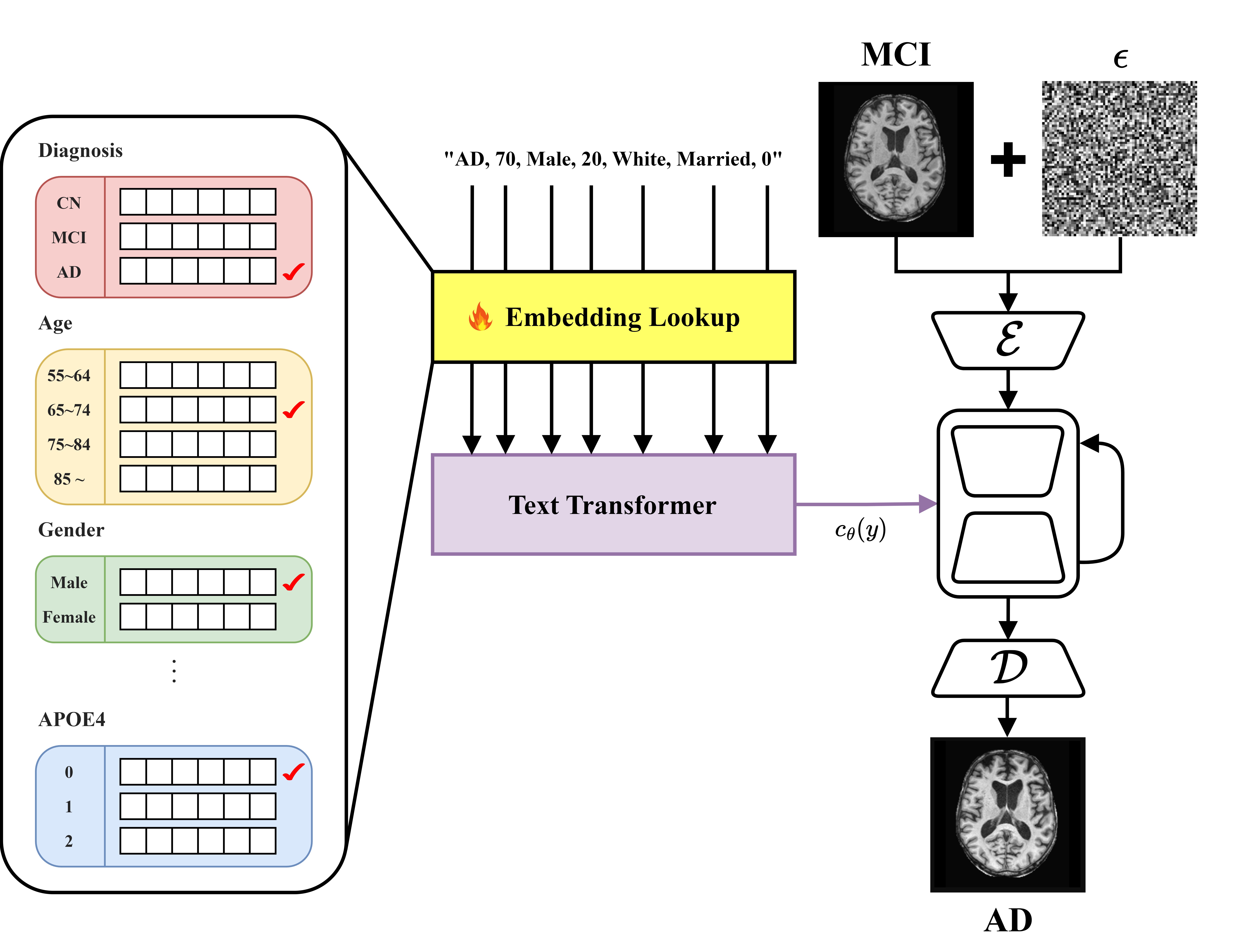
- This study developed a diffusion model that integrates clinical and genetic data to generate T1-weighted MRI images corresponding to various stages of Alzheimer’s disease (CN, MCI, AD).
- Utilizing a pre-trained text-to-image model, Stable Diffusion [1], this approach generates medical images and overcomes challenges associated with medical terminology and domain shifts through textual inversion [2] and fine-tuning techniques.
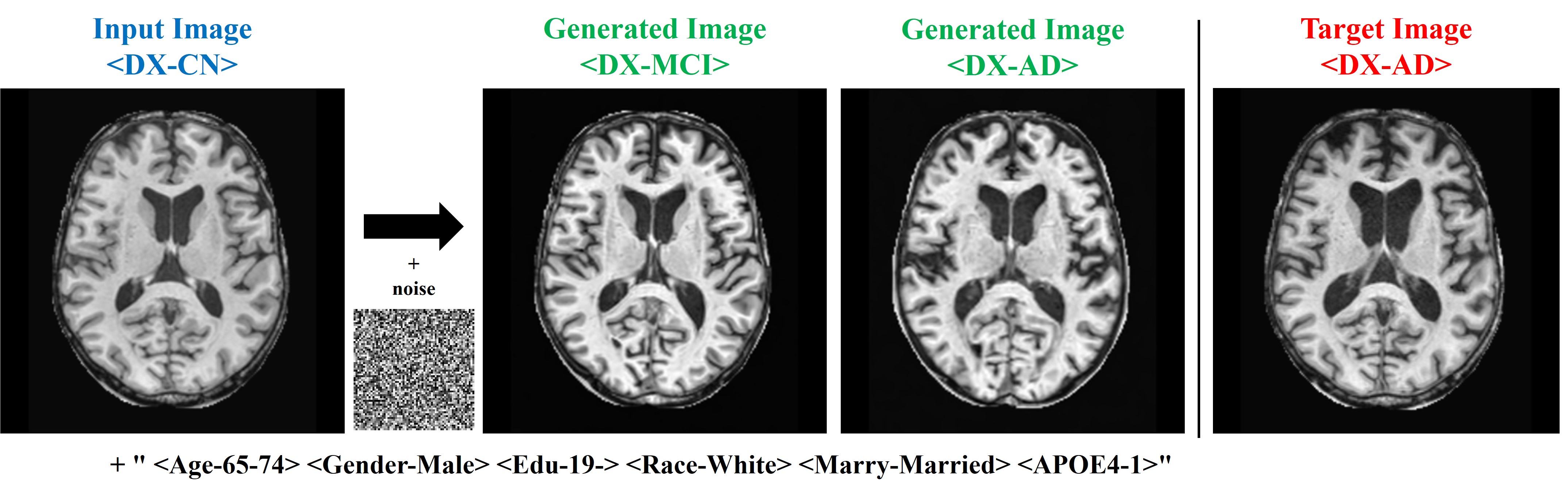
- Experimental results confirm that this model can effectively predict structural brain changes associated with the progression of Alzheimer’s disease by comparing generated MRIs with actual MRIs.
- This capability is anticipated to make significant contributions to early diagnosis and progression prediction.
In this study, we utilized 6,355 T1-weighted MRI datasets. Of these, 308 datasets with confirmed disease progression were designated for evaluation, while the remainder were used for training. The T1-weighted MRI data were standardized through N4ITK bias correction and rigid registration with the MNI152 T1 1mm atlas [4-7]. Subsequently, brain extraction was performed to remove non-brain tissues. The central axial slice of the processed images was extracted for model training and evaluation.
Baseline was defined as the difference from the input MRI before the transition.
| FID ↓ | PSNR ↑ | SSIM ↑ | LPIPS ↑ | |
|---|---|---|---|---|
| Baseline | 0.0017 | 24.5068 | 0.9309 | 0.0445 |
| Ours | 0.0105 | 23.5541 | 0.8953 | 0.0730 |
Despite limitations in predicting fine structures due to noise addition, the improvement in LPIPS score demonstrates the model's ability to generate images aligned with human visual perception. This indicates that the model can effectively predict structural brain changes associated with Alzheimer's progression, even in the absence of long-term follow-up data.
- [1] Rombach, Robin, et al. "High-resolution image synthesis with latent diffusion models." Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp. 10684-10695, 2022.
- [2] Gal, Rinon, et al. "An image is worth one word: Personalizing text-to-image generation using textual inversion." arXiv preprint arXiv:2208.01618, 2022.
- [3] Weiner, Michael W., et al. "The Alzheimer's Disease Neuroimaging Initiative 3: Continued innovation for clinical trial improvement." Alzheimer's & Dementia, Vol. 13, No. 5, pp. 561-571, May. 2017.
- [4] Tustison, Nicholas J., et al. "N4ITK: improved N3 bias correction." IEEE transactions on medical imaging, Vol. 29, No. 6, pp. 1310-1320, April. 2010.
- [5] Fonov, Vladimir, et al. "Unbiased average age-appropriate atlases for pediatric studies." Neuroimage, Vol. 54, No. 1, pp. 313-327, January. 2011.
- [6] Fonov, Vladimir S., et al. "Unbiased nonlinear average age-appropriate brain templates from birth to adulthood." NeuroImage, Vol. 47, pp. S102, July. 2009.
- [7] Collins, D. Louis, et al. "ANIMAL+ INSECT: improved cortical structure segmentation." Information Processing in Medical Imaging: 16th International Conference, IPMI’99 Visegrád, Hungary, pp. 210-223, June 28–July 2, 1999.