Project for Computational Fluid Dynamics Lab course.
Group 6: Nathan Brei, Irving Cabrera, Natalia Saiapova.
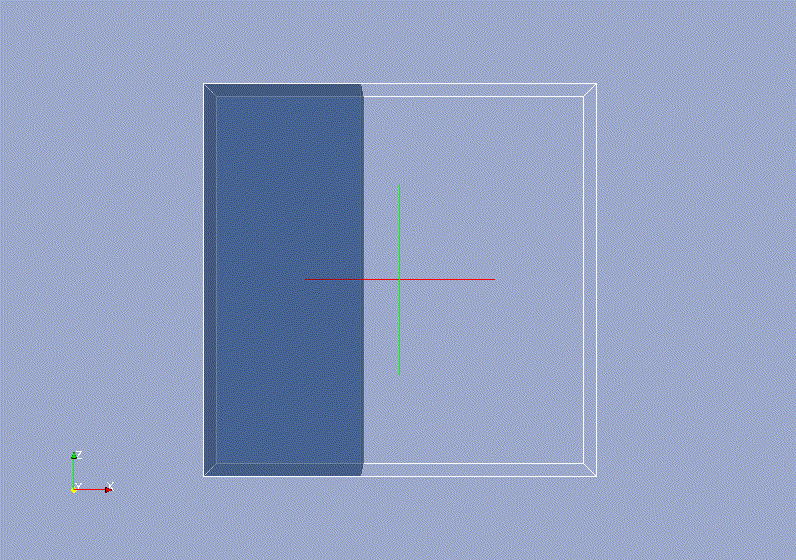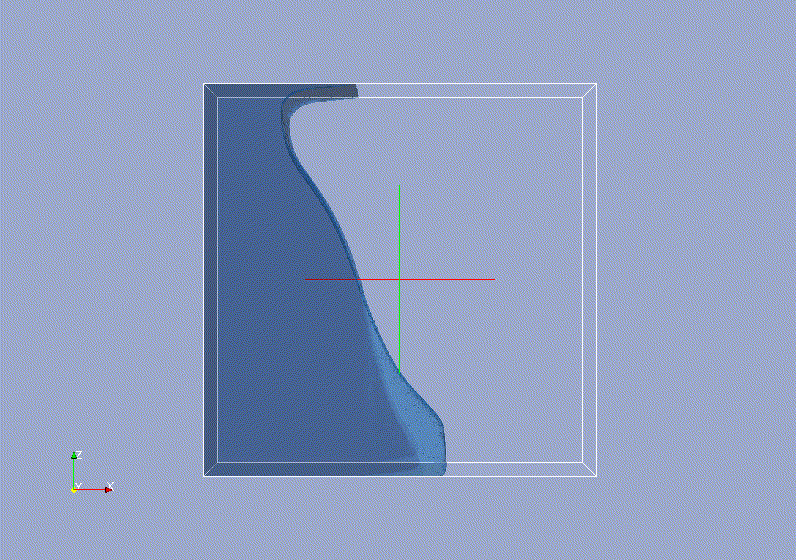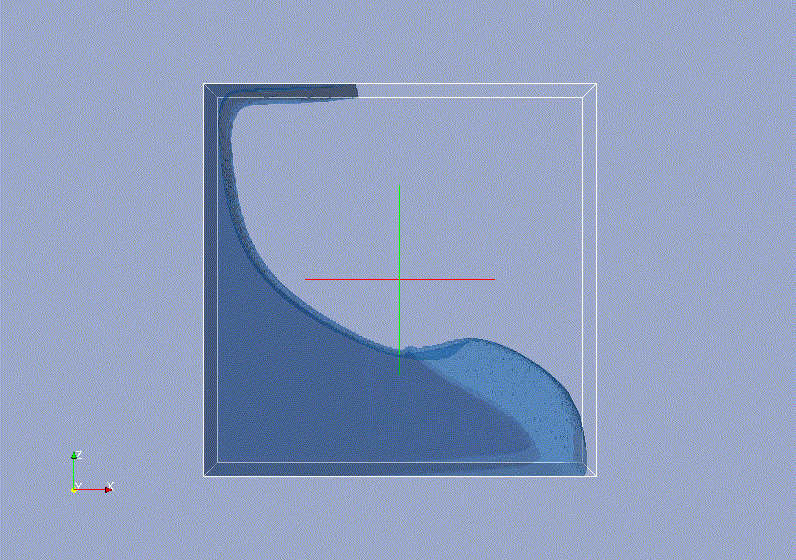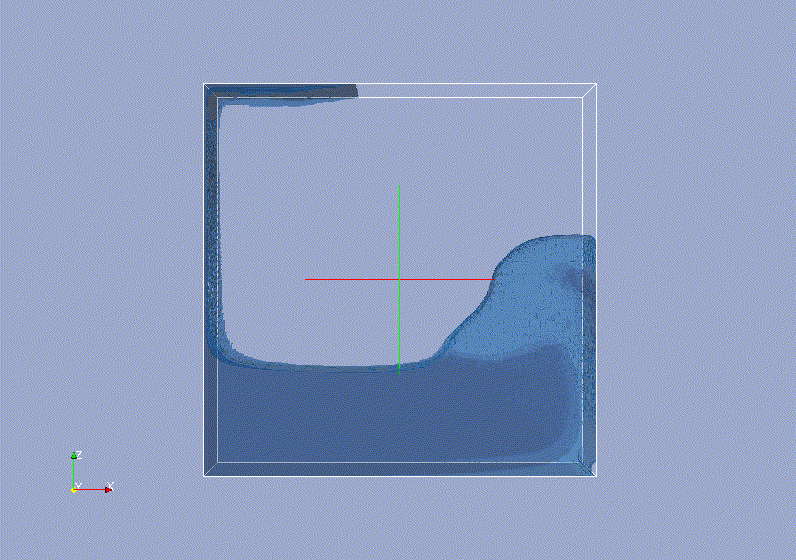

Requirements: UNIX based OS.
Use make command to compile:
>> make
There is debug mode available. This mode includes additional checks for velocity and density, computes total mass in the domain and checks that there is no FLUID cell which is neighboring with a GAS cell directly. Also, it calculates the time spent on the following parts: streaming, collision, flag update and boundary treatment.
>> make debug
To run the program use
>> ./lbsim <exampleName> <number of threads>
For each example it is required to have <exampleName>.dat and <exampleName>.pgm files. They have to be placed in the examples directory.
<number of threads> specifies how many threads will be used by OpenMP.
The vtk-output directory contains vtk output files. If thare is no such directory it will be created.
We added a few new fields to the input.dat file:
| Parameter | Description |
|---|---|
| radius | Radius of a droplet |
| exchange_factor | Factor to increase speed of a mass exchnge (mainly to get more beautiful videos) |
| forces_x | Gravity force in the x direction |
| forces_y | Gravity force in the y direction |
| forces_z | Gravity force in the z direction |
First of all, we added 2 new cell types: FLUID and INTERFACE.
Here is table representing cell types, which should be used in <exampleName>.pgm file:
| Cell type | Value | Cell type | Value |
|---|---|---|---|
| FLUID | 0 | PRESSURE_IN | 5 |
| INTERFACE | 1 | OBSTACLE | 6 |
| MOVING_WALL | 2 | FREESLIP | 7 |
| INFLOW | 3 | NOSLIP | 8 |
| OUTFLOW | 4 | GAS | 9 |
In the checks.c one can find three check functions:
check_in_rankchecks that everyFLUIDcell has a density between0.9and1.1and norm of the velocity vector is less thensqrt(3) * C_S;check_flagschecks that noFLUIDcell has aGASneighbor;check_masscompute total mass in the domain (FLUIDandINTERFACEcells).
This checks will be activated in debug mode and be executed every time when we write output file.
In collision step additional term for external forces was added.
Related function in collision.c: computeExternal(int i, float density, float * extForces), where extForces is a force vector specified in input file.
For every cell we store 2 additional float values: mass and fluid fraction. We update mass field during streaming step. Fluid fraction field is updated during collide step.
To be able get more fast we introduced exchange_factor in an input file. By this factor we increase mass exchange between cells.
In the streaming step was added reconstruction from GAS cells. Streaming works normally for all FLUID cells amd for INTERFACE cells we are checking whether a neighbor is GAS. If yes, we are using outflow boundary formula in this direction.
From the mass fraction for each cell we determine whether that cell emptied or filled. We track these cells using two arrays, emptiedCells and filledCells. We perform the filling and emptying in separate phases, performFill() and performEmpty(). These phases are asymmetric in order to handle the case of a cell filling adjacent to a cell emptying. If this happens, the 'filling' operation cancels the neighbor's 'emptying' operation and the neighboring cell is stricken from emptiedCells.
Both phases maintain the loop invariant of a contiguous interface layer. When an INTERFACE cell is converted to FLUID, all GAS neighbors are converted to INTERFACE. Correspondingly, when an INTERFACE is converted to GAS, all FLUID cells are converted to INTERFACE.
We have added pragma omp parallel for to streaming, collision, treatment boundaries and update flags.
At the beginning our main bottleneck was computeFeq function which took approximately 42% percent of the whole time. We splitted the Q-loop in this function in a way that compiler was able to vectorize it, then we got rid of all divisions and replaced them by corresponding multiplications. Then we changed types of all double fields to float. It gave us aproximately 2x speedup.