ProkBERT is an advanced genomic language model specifically designed for microbiome analysis. This repository contains the ProkBERT package and utilities, as well as the LCA tokenizer and model definitions.
- Introduction
- Features
- Installation
- Applications
- Quick start
- Tutorials and examples
- ProkBERT's results
- Citing this work
The ProkBERT model family is a transformer-based, encoder-only architecture based on BERT. Built on transfer learning and self-supervised methodologies, ProkBERT models capitalize on the abundant available data, demonstrating adaptability across diverse scenarios. The models’ learned representations align with established biological understanding, shedding light on phylogenetic relationships. With the novel Local Context-Aware (LCA) tokenization, the ProkBERT family overcomes the context size limitations of traditional transformer models without sacrificing performance or the information-rich local context. In bioinformatics tasks like promoter prediction and phage identification, ProkBERT models excel. For promoter predictions, the best-performing model achieved an MCC of 0.74 for E. coli and 0.62 in mixed-species contexts. In phage identification, they all consistently outperformed tools like VirSorter2 and DeepVirFinder, registering an MCC of 0.85. Compact yet powerful, the ProkBERT models are efficient, generalizable, and swift.
- Tailored to microbes.
- Local Context-Aware (LCA) tokenization for better genomic sequence understanding.
- Pre-trained models available for immediate use and fine-tuning.
- High performance in various bioinformatics tasks.
- Facilitation of both supervised and unsupervised learning.
The recommended way to install ProkBERT is through pip, which will handle most dependencies automatically:
pip install prokbertProkBERT is also available as a conda package from the Bioconda channel. To install it using conda, run:
conda install prokbert -c biocondaBefore using the ProkBERT container with GPU support, make sure you have the following installed on your system:
- Python (3.10 or later)
- Docker (required if you plan to use the Docker image)
- NVIDIA Docker (required if you intend to use Docker with GPU support)
To pull and run the ProkBERT Docker image, use:
docker pull obalasz/prokbertTo run the container with GPU support, use:
docker run --gpus all -it --rm -v $(pwd):/app obalasz/prokbert python /app/finetuning.py --helpThis command runs the ProkBERT Docker container with GPU support, mounts the current directory to the container (allowing access to your local files), and displays the help message for the finetuning.py script.
For users who prefer Singularity (now also known as Apptainer), a Singularity (Apptainer) container definition is provided. Please refer to the official documentation for instructions on building and running Singularity containers. A prebuilt container is available on Zenodo: https://zenodo.org/records/10659030.
Building the Singularity container:
apptainer build prokbert.sif prokbert.defTo pull directly from Docker Hub and convert to a Singularity image file:
singularity pull prokbert.sif docker://obalasz/prokbertOnce you have your .sif file, you can run ProkBERT with the following command:
singularity run --nv prokbert.sif python /opt/prokbert/examples/finetuning.py --helpThe --nv flag enables NVIDIA GPU support, assuming your system has NVIDIA drivers and CUDA installed. Remove this flag if you're running on a CPU-only system.
You can also shell into the container or execute specific commands as follows: Shell into the container:
singularity shell --nv prokbert.sifNote: If you encounter any problems, please do not hesitate to contact us or open an issue. My email address is obalasz@gmail.com.
ProkBERT has been validated in several key genomic tasks, including:
- Learning meaningful repreresentation for seqeuences (zero-shot capibility)
- Accurate bacterial promoter prediction.
- Detailed phage sequence analysis within complex microbiome datasets.
Our models and datasets are avaialble on the hugginface page. The models are easy to use with the transformers package. We provide examples and descriptions as notebooks in the next chapter and some example scsripts regarging how to preprocess your sequence data and how to finetune the available models. The examples are available in the example folder of this repository.
To load the model from Hugging Face:
import torch
from transformers import AutoTokenizer, AutoModel
from prokbert.prokbert_tokenizer import ProkBERTTokenizer
tokenizer = ProkBERTTokenizer(tokenization_params={'kmer' : 6, 'shift' : 1})
model = AutoModel.from_pretrained("nerualbioinfo/prokbert-mini", trust_remote_code=True)
segment = "TATGTAACATAATGCGACCAATAATCGTAATGAATATGAGAAGTGTGATATTATAACATTTCATGACTACTGCAAGACTAA"
inputs = tokenizer(segment)['input_ids']
encoded_sequence = tokenizer.batch_encode_plus([segment])
# Converting the ids to tensors of int64
encoded_sequence = {key: torch.tensor(val, dtype=torch.int64) for key, val in encoded_sequence.items()}
model(**encoded_sequence)For examples of how to preprocess the raw sequence data, which are frequently stored in fasta format: Examples:
- Segmentation (sequence preprocessing): colab link
- Tokenization colab link
- Preprocessing for pretraining
An example of how to visualize the genomic features of ESKAPE pathogens. More description about the dataset is available on Hugging Face Example:
- ESKAPE pathogen genomic features: colab link
Here we provide an example of a practical transfer learning task. It is formulated as a binary classification. We provide a notebook for presenting the basic concepts and a command line script as a template. Examples:
- Finetuning for promoter identification task: colab link
- Python script for the finetuning: link
Usage example:
git clone https://github.com/nbrg-ppcu/prokbert
cd examples
python finetuning.py \
--model_name neuralbioinfo/prokbert-mini \
--ftmodel mini_promoter \
--model_outputpath finetuning_outputs \
--num_train_epochs 1 \
--per_device_train_batch_size 128 For practical applications or for larger training tasks we recommend using the Distributed DataParallel.
In this section, we provide a practical example for evaluating finetuned ProkBERT models. This is crucial for understanding the model's performance on specific tasks, such as promoter prediction or phage identification.
We have prepared a detailed example that guides you through the process of evaluating our finetuned models. This includes loading the model, preparing the data, running the inference, and interpreting the results.
Example:
Here you can find an example of pretraining ProkBERT from scratch. Pretraining is an essential step, allowing the model to learn the underlying patterns before being fine-tuned for downstream tasks. All of the pretrained models are available on Hugging Face.
| Model | k-mer | Shift | Hugging Face URL |
|---|---|---|---|
| ProkBERT-mini | 6 | 1 | Link |
| ProkBERT-mini-c | 1 | 1 | Link |
| ProkBERT-mini-long | 6 | 2 | Link |
It is a good practice to preprocess larger sequence sets (>100MB) in advance. Below is an example script for achieving this by running on multiple fasta files:
Clone the ProkBERT repository and navigate to the examples directory. Then, preprocess the example data into a format suitable for training. This involves converting sequences into k-mer representations. Run the following commands:
git clone https://github.com/nbrg-ppcu/prokbert
cd prokbert/examples
python prokbert_seqpreprocess.py \
--kmer 6 \
--shift 1 \
--fasta_file_dir ../src/prokbert/data/pretraining \
--out ../src/prokbert/data/preprocessed/pretraining_k6s1.h5Parameters:
--kmer: The size of the k-mer (number of bases) to use for sequence encoding.--shift: The shift size for sliding the k-mer window across sequences.--fasta_file_dir: Directory containing your FASTA files for pretraining.--out: Output file path for the preprocessed data in HDF5 format.
Use the preprocessed HDF file as input for pretraining. Execute the commands below:
python prokbert_pretrain.py \
--kmer 6 \
--shift 1 \
--dataset_path ../src/prokbert/data/preprocessed/pretraining_k6s1.h5 \
--model_name prokbert_k6s1 \
--output_dir ./tmppretraining \
--model_outputpath ./tmppretrainingParameters:
--model_name: Name for the model configuration to be used or saved.--output_dir: Directory where the training logs and temporary files will be saved.--model_outputpath: Path where the final trained model should be saved.
ProkBERT is a novel genomic language model family designed for microbiome studies, pretrained on extensive genomic datasets from the NCBI RefSeq database. The pretraining covered a diverse range of organisms, utilizing over 206.65 billion tokens from various genomes, encompassing bacteria, viruses, archaea, and fungi. The final dataset included 976,878 unique contigs from 17,178 assemblies, representing 3,882 distinct genera. Detailed methodology and tokenization strategies are elaborated in our paper.
ProkBERT's embeddings provide insights into genomic structures and relationships, distinguishing between different genomic elements. The models showcase strong generalization abilities, aligning with established biological principles.
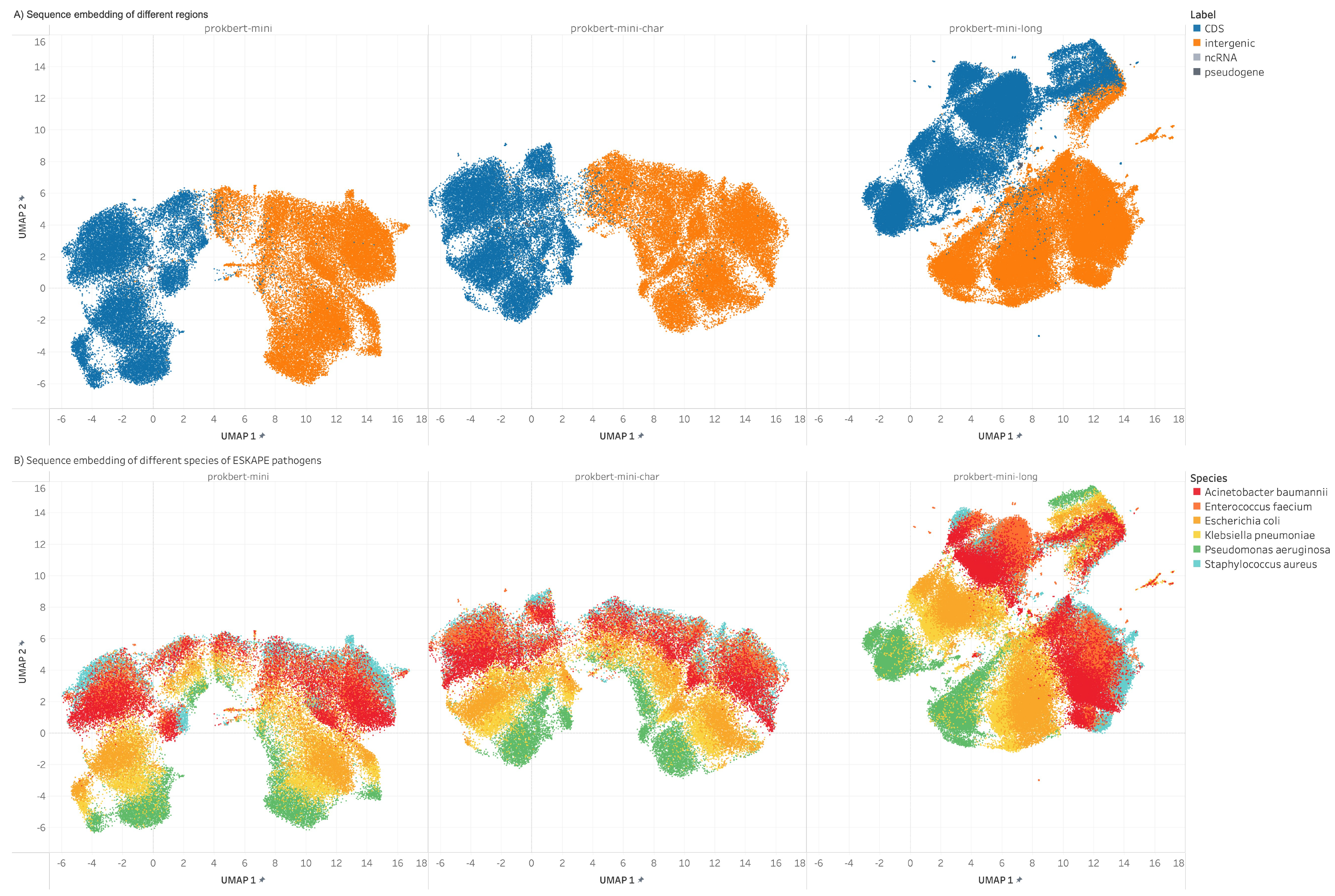 Figure 1: UMAP embeddings of genomic segment representations, highlighting the distinction between genomic features.
Figure 1: UMAP embeddings of genomic segment representations, highlighting the distinction between genomic features.
ProkBERT models have been applied to identify bacterial promoters, demonstrating superior accuracy and robustness compared to existing tools.
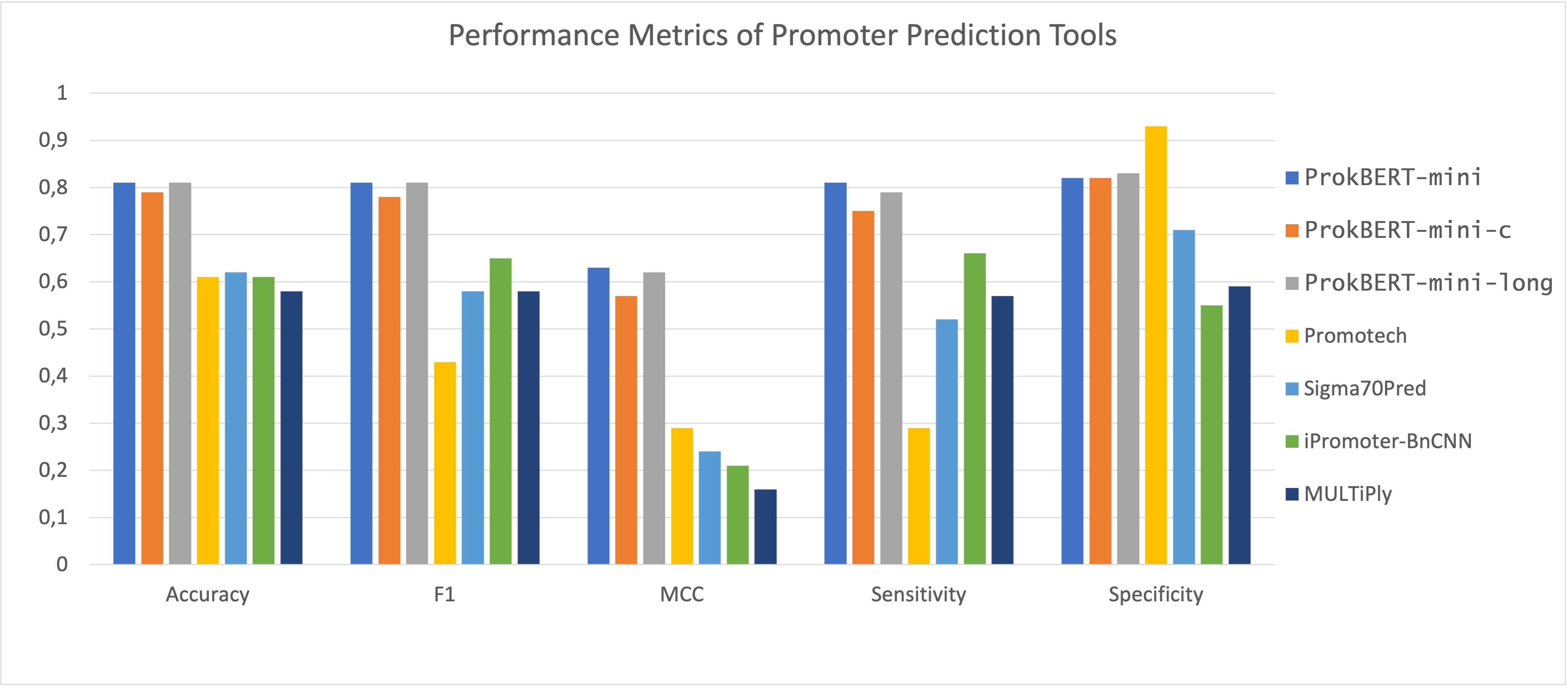 Figure 2: Performance metrics of ProkBERT in promoter prediction.
Figure 2: Performance metrics of ProkBERT in promoter prediction.
ProkBERT models demonstrated high performance with an impressive accuracy and MCC (Matthews Correlation Coefficient) of 0.87 and 0.74, outperforming other established tools such as CNNProm and iPro70-FMWin. This highlights ProkBERT's effectiveness in correctly identifying both promoters and non-promoters, with consistent results across various model variants. The evaluation also included a comparative analysis with newer tools like Promotech and iPromoter-BnCNN.
In phage sequence analysis, ProkBERT outperforms traditional methods, proving its efficacy in identifying phage sequences within complex genomic data.
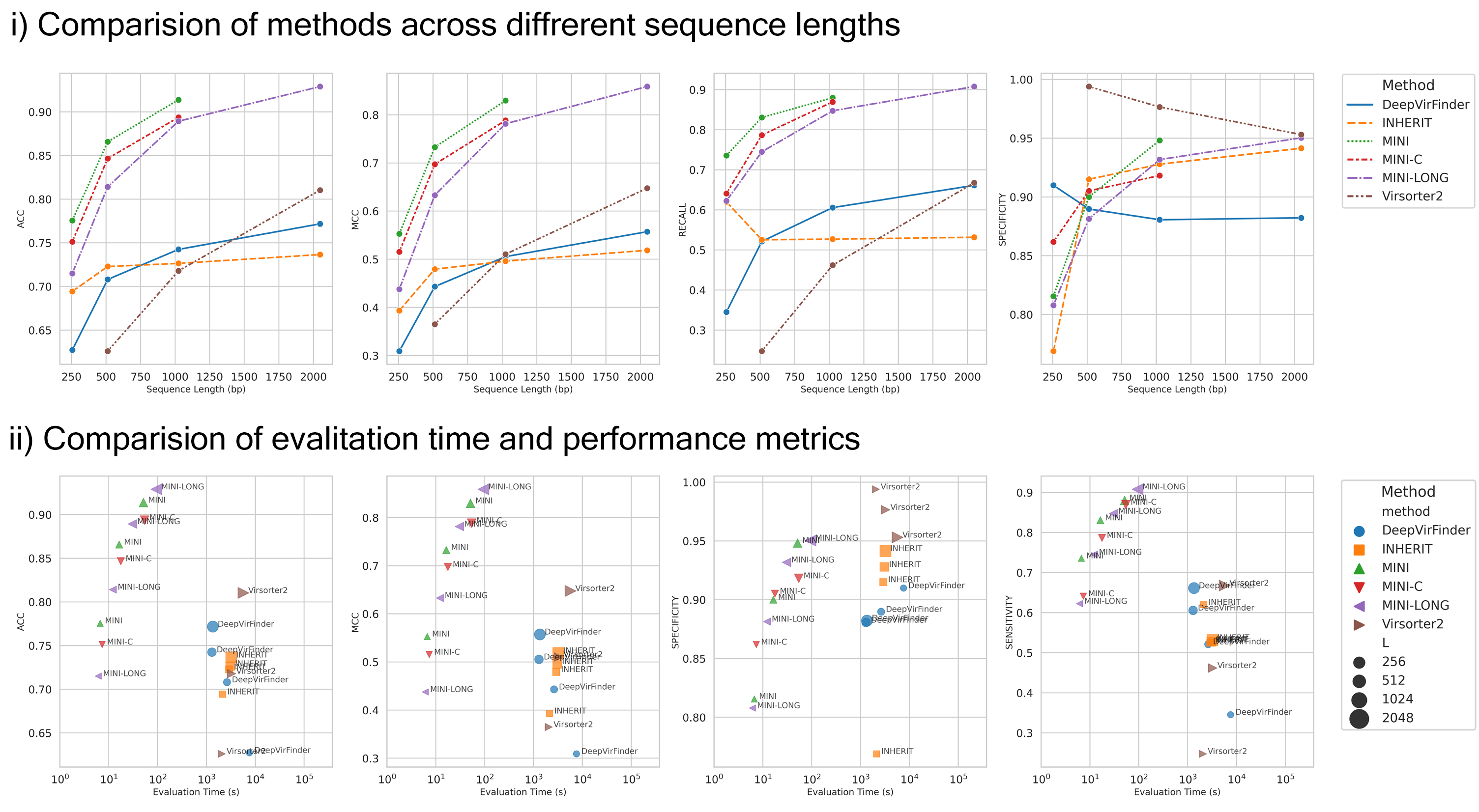 Figure 3: Comparative analysis showcasing ProkBERT's performance in phage prediction.
Figure 3: Comparative analysis showcasing ProkBERT's performance in phage prediction.
Our evaluations demonstrate the performance of ProkBERT in classifying phage sequences. It achieves high sensitivity and specificity even in challenging cases where available sequence information is limited. However, this exercise also highlights an inherent limitation of ProkBERT, the restricted context window size. In comparative benchmarks with varying short sequence lengths, ProkBERT consistently surpassed established tools like VirSorter2 and DeepVirFinder
| Model Name | k-mer | Shift | Hugging Face URL |
|---|---|---|---|
neuralbioinfo/prokbert-mini |
6 | 1 | Link |
neuralbioinfo/prokbert-mini-long |
6 | 2 | Link |
neuralbioinfo/prokbert-mini-c |
1 | 1 | Link |
| Model Name | k-mer | Shift | Hugging Face URL |
|---|---|---|---|
neuralbioinfo/prokbert-mini-promoter |
6 | 1 | Link |
neuralbioinfo/prokbert-mini-long-promoter |
6 | 2 | Link |
neuralbioinfo/prokbert-mini-c-promoter |
1 | 1 | Link |
| Model Name | k-mer | Shift | Hugging Face URL |
|---|---|---|---|
neuralbioinfo/prokbert-mini-phage |
6 | 1 | Link |
neuralbioinfo/prokbert-mini-long-phage |
6 | 2 | Link |
neuralbioinfo/prokbert-mini-c-phage |
1 | 1 | Link |
| Dataset Name | Hugging Face URL |
|---|---|
neuralbioinfo/ESKAPE-genomic-features |
Link |
neuralbioinfo/phage-test-10k |
Link |
neuralbioinfo/bacterial_promoters |
Link |
neuralbioinfo/ESKAPE-masking |
Link |
If you use the code or data in this package, please cite:
@Article{ProkBERT2024,
author = {Ligeti, Balázs and Szepesi-Nagy, István and Bodnár, Babett and Ligeti-Nagy, Noémi and Juhász, János},
journal = {Frontiers in Microbiology},
title = {{ProkBERT} family: genomic language models for microbiome applications},
year = {2024},
volume = {14},
URL={https://www.frontiersin.org/articles/10.3389/fmicb.2023.1331233},
DOI={10.3389/fmicb.2023.1331233},
ISSN={1664-302X}
}