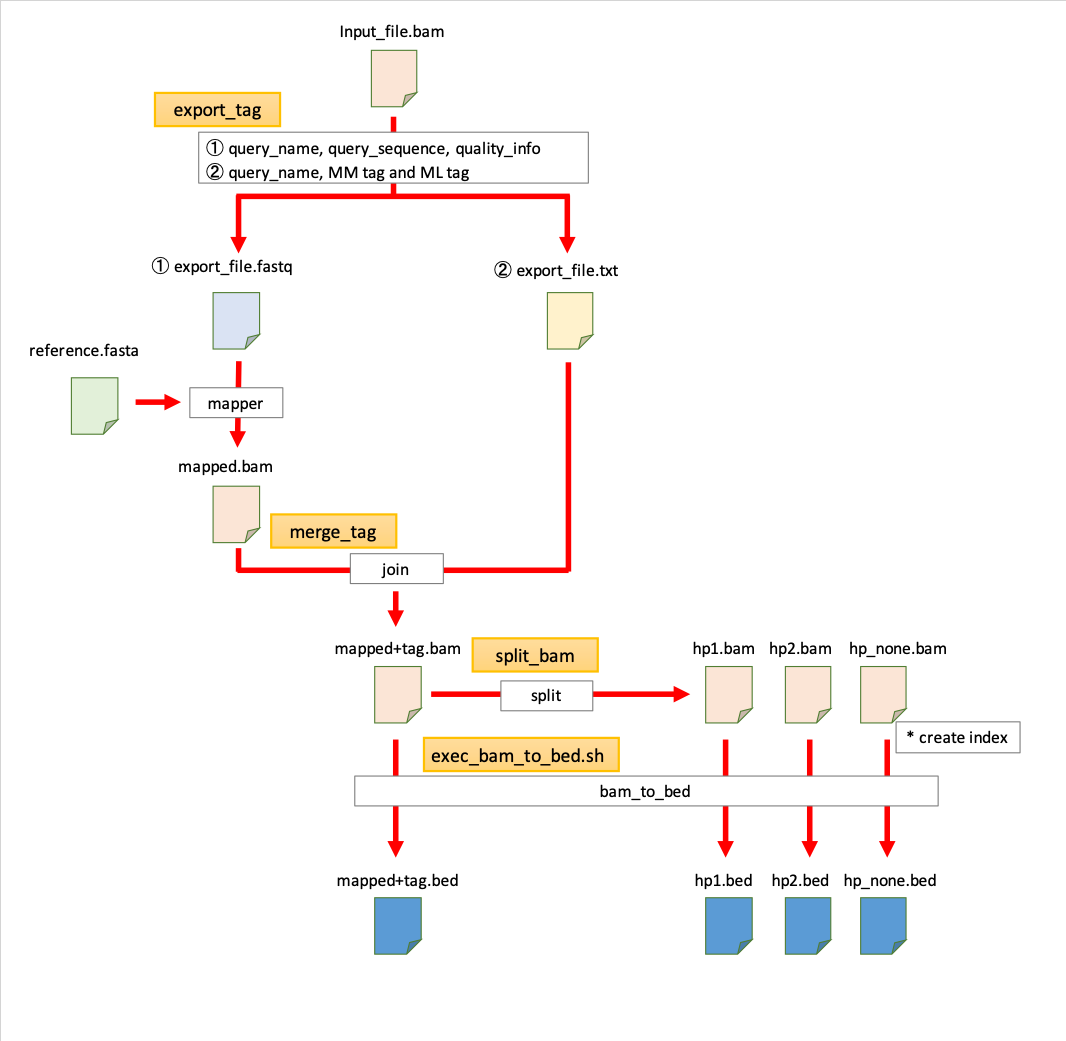
This script can output the fastq file and mm/ml tag information separately from the bam file. After mapping the output fastq file with any mapper, the process of merging mm/ml tag information is executed.
- Docker
git clone https://github.com/ncc-ccat-gap/ModbamETLTools.git
docker build ModbamETLTools/Dockerfile -t modbametltools:0.2.0
docker run -it modbametltools:0.2.0 /bin/bash
- When starting the container, please mount the input/output destination according to the environment.
- If you also have a singularity environment, you can also create a .sif to use.
singularity build modbamETLtools-0.2.0.sif docker-daemon://modbametltools:0.2.0
- It can be used even if you do not have a Docker environment.
- In that case, please set up the following environment
- Python 3.8.x
- pysam 0.20.0
- modbam2bed v0.6.3
git clone https://github.com/ncc-ccat-gap/ModbamETLTools.git
cd ModbamETLTools
python setup.py install
- <exec_bam_to_bed.sh> is a script created with the assumption that it will run in a Docker environment.
- The modbam2bed path in the <exec_bam_to_bed.sh> file should be rewritten according to your environment
export_tag -i <input_bam_file> [-o output_file_path] [--no_fastq]
merge_tag -i <input_bam_file> -t <input_mm/ml_tag_file> [-o output_file_path] [--no_sort] [--threads <int>]
split_bam -i <input_bam_file> [-o output_file_path]
cd ModbamETLTools/scripts/etl_tools
bash exec_bam_to_bed.sh <input_bam_file> <fasta_file> <threads>
- Example: qsub option for thread 8 is as follows
- def_slot 4
- s_vmem 4G
export_tag -i /home/user/sample1.bam
- If option '-o' is omitted, the output is current.
- If '--no_fastq' is given as an argument, the .fastq file is not output
- sample1_export.fastq
- sample1_export_taginfo.txt
merge_tag -i /home/user/sample2.bam -t /home/user/sample1_export_taginfo.txt
- If option '-o' is omitted, the output is current.
- If '--no_sort' is given as an argument, the sorting process is skipped.
- You can specify the number of cores to process by giving '--threads' as an argument.(default:8)
- sample2_merge_tags.bam
- sample2_merge_tags_sorted.bam
- sample2_merge_tags_sorted.bam.bai
split_bam -i /home/user/sample2_merge_tags_sorted.bam
- If option '-o' is omitted, the output is current.
- sample2_merge_HP_none.bam
- sample2_merge_HP2.bam
- sample2_merge_HP1.bam
- sample2_merge_HP1.bam.bai
- sample2_merge_HP2.bam.bai
- sample2_merge_HP_none.bam.bai
cd ModbamETLTools/scripts/etl_tools
bash exec_bam_to_bed.sh /home/user/sample2_merge_HP_none.bam /home/user/reference.fasta 8
-
sample2_merge_HP_none_mod-counts.cpg.acc.bed
-
sample2_merge_HP_none.bed
- '*-counts.cpg.acc.bed' : strands are aggregated