This repository contains the implementation of some of the ideas in our paper https://arxiv.org/pdf/2103.04470.pdf.
The implementation below is via Matlab's parfor.
The software was developed by Mario Gomez and Facundo Memoli. We also use functions written by Misha Belkin (L2_distance.m), Vin de Silva (px_fps.m), Uli Bauer (Ripser: https://github.com/Ripser/ripser), Masoud Alipur (emd_mex.m), and Michael Kerber, Dmitriy Morozov & Arnur Nigmetov (bottleneck distance with Hera). We have also benefited from Chris Tralie's Matlab wrapper for Ripser (https://github.com/ctralie/Math412S2017).
- This code was developed in Matlab version R2019a. It requires the Parallel Computing Toolbox (https://www.mathworks.com/products/parallel-computing.html).
- From a terminal go to the ripser folder and run
make ripser-coeff.
The scripts in the base directory (i.e. Persistence_parallel_dX.m and Persistence_parallel_Ripser.m) compute the principal persistence sets .mat file. The program will load this matrix and sample a large number of
We have included a file dX_example_circle.mat with 1000 points sampled uniformly at random from the unit circle. As long as you have both Ripser and the Parallel Computing Toolbox installed, both Persistence_parallel_Ripser.m and Persistence_parallel_dX.m should run out of the box. You can further personalize the scripts as described in the Usage section.
You need to set the following parameters:
- Store a distance matrix named
dXin a .mat file. In order to guarantee meaningful results, the matrix should be square, symmetric, have diagonal 0, and satisfy the triangle inequality.- Keep in mind that a reasonable size for
dXis at most 5000x5000, depending on your system.
- Keep in mind that a reasonable size for
- Select the script that you want to use (Persistence_parallel_Ripser.m or Persistence_parallel_dX.m). Open the file to set the following book-keeping variables:
-
filename_metric_space: Name (and path) of the .mat file containing the distance matrix. -
results_file: The name of the file where the results will be stored. -
save_to_file: Boolean flag to decide if you want to save the output to results_file.mat or if you only want to keep it in Matlab's workspace. -
temp_file: The ending of the file where the temporary results will be stored. The files are named checkpoints_temp_file_#.mat, where # runs from 1 to the number of cores utilized. This variable is usually not edited, unless you are running several copies of the script at the same time. -
CoresRequested: The number of cores that you want to use. The maximum is the number of physical cores (not logical cores) available in your system. You can setCoresRequested = 0to use the default chosen by Matlab.- Matlab can only work with physical cores. See https://www.mathworks.com/matlabcentral/answers/80129-definitive-answer-for-hyperthreading-and-the-parallel-computing-toolbox-pct#answer_89845 for a more detailed explanation. In short, logical cores require hyperthreading, but this negatively impacts Matlab's performance more often than not.
-
- Set the simulation parameters:
-
dim: The dimension of homology that you want to calculate. The program will automatically setn=2*dim+2to calculate the principal persistence set. -
nReps: Number of$n$ -point samples to take from$d_X$ .
-
- Once you've decided on the above parameters, run your selected script from the Matlab command window. This will produce a .mat file containing the results.
The script produces a graph of
-
confs: Matrix of size[n, nReps]. Each columnI = confs(:,i)is the set of indices a set of$n$ points sampled from$X$ . -
dms: Array of size[n, n, nReps]. Each pagedm = dms(:,:,i)is the distance matrix of a sample of$n$ points from$X$ . It is obtained fromdXandconfsby settingI=confs(:,i)anddm=dX(I,I). -
bd_times: An array of size[nReps, 2]with the persistence diagrams of the samples. The first columnbd_times(:,1)contains the birth times and the second,bd_times(:,2), the death times. A rowbd_times(i,:)will be non-zero if, and only if, the configuration with distance matrixdms(:,:,i)produced persistence homology.
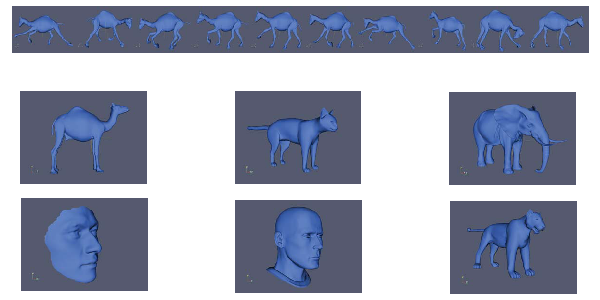
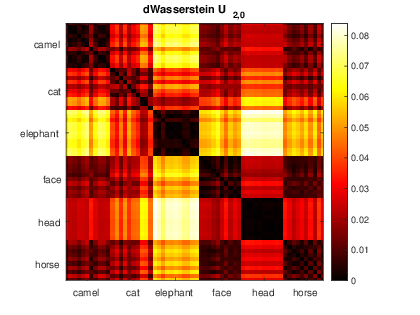
We refer the user to Section 4.3.1 of our paper for the description of the experiment. We used the database from the paper ''Deformation Transfer for Triangle Meshes'' by Robert W. Sumner and Jovan Popović. As of the time of writing, both the database and the paper can be found here: https://people.csail.mit.edu/sumner/research/deftransfer/.
To use this code, download the database from the link above. Place the folders with the meshes in the models directory. To reproduce the experiments in our paper, you should have the following subfolders: camel-poses, cat-poses, elephant-poses, face-poses, head-poses, and horse_poses. Then, run the scripts in the Shape_Analysis folder in order (run S1_Setup, then S2_persistence_Sets.m, and so on). The order is set up so that, if you change parameters in one script, you don't have to re-run every script before it. These parameters are:
-
S1_setup.m:
-
nBins0andnBins1are the number of points used to approximate$U_{2k+2,k}^\mathrm{VR}(G_i)$ for$k=0$ and$k>0$ , respectively. Be aware that the number of points is only guaranteed to be close tonBins0andnBins1. Choose these numbers so thatemd_mexcan compute the Wasserstein distance between vectors with that size. -
NFPS: Number of points in the graphs$G_i$ . ChooseNFPSso that MATLAB can store a matrix of size[NFPS,NFPS].
-
-
S2_Persistence_Sets.m:
-
Ks: Vector with the dimensionskto compute the persistence sets$D_{2k+2,k}$ . -
nRepsK: Vector of the same size asKs. The entrynRepsK(idx)is the number of samples to take to compute$D_{2k+2,k}$ fork=Ks(idx). Note: This value is unused whenk=0, butnRepsKneeds a placeholder value.
-
-
S3_Persistence_Diagrams.m:
-
maxHomDim: We compute the persistence diagrams from dimension 0 up tomaxHomDim. -
nL: We usually can't compute the persistence diagram of the whole set$G_i$ . Instead, we operate on a subset of sizenL. -
s_edges: We compute persistence diagrams up to a thresholdtchosen so that the 1-skeleton of the Vietoris-Rips complex contains a certain percentage of the total number of edges. That percentage is0 < s_edges < 1.
-
-
S4_NN_Classification.m:
-
nTests: The number of times that the 1-nearest neighbor experiment will be repeated.
-
-
S5_NN_Classification_dWmax.m:
-
nTests: Same as in S4_NN_Classification.m. - Line 88: You can modify the initial guess
w0here.
-
We include a folder with further experiments.
The script Persistence_Single_Ellipse.m computes the principal persistence set of an ellipse in k, nReps, save_to_file, and results_file as above. This code is not parallelized. Additionally, you can modify the length of the semiaxes a and b. The variable confs now has size [n, 2, nReps], and each page confs(:,:,i) is an a and b.
 a=1.2, b=1.0.png?raw=true "D41_Ellipse")
An image similar to the above can be produced by running Persistence_Single_Ellipse.m with a=1.2, b=1, k=1, and nReps=10^6.
These scripts compute principal persistence sets of metric graphs. For the script Persistence_Metric_Graph.m, the input is a weighted adjacency matrix stored in the variable A. We include several examples (commented in the file). Persistence_Metric_Graph_Collection.m can operate several graphs at once. The input is a set of weighted adjacency matrices, each separated by an empty line, stored in a text file in the Other_Examples/Graphs/data/ folder. The folder includes several examples.
Similar to the main script, the variables that can be modified in these scripts are k, nReps, save_to_file, and results_file. If you set save_to_file=true in Persistence_Metric_Graph_Collection.m, the script will save the graphs of the persistence sets to the 'Results/' folder instead of showing them on screen.
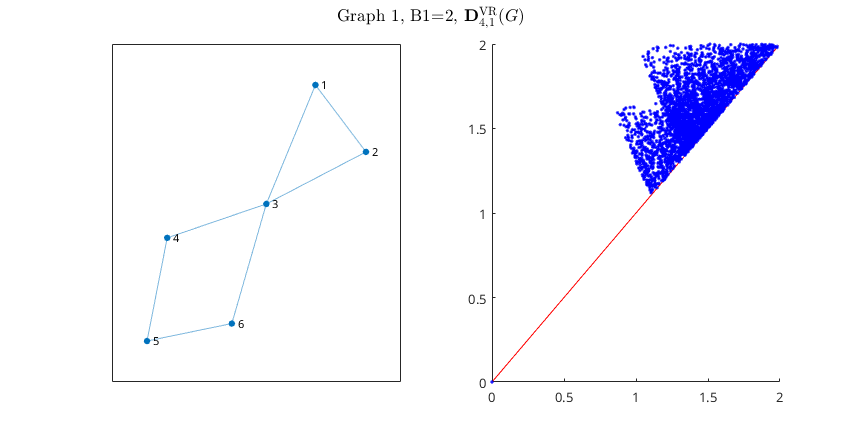 The image above was obtained by running Persistence_Metric_Graph.m with
The image above was obtained by running Persistence_Metric_Graph.m with k=1 and nReps=10^5. Among the adjacency matrices in the file, we uncommented the one titled "Wedge of two circles of different lengths".
We have observed that sampling 6-point configurations from k and set n=2k+2. To approximate nReps configurations of n points from nSteps steps as follows. It starts with a configuration
The variables that can be modified in this script are:
-
d: The dimension of the sphere$\mathbb{S}^d$ . -
sigma: The standard deviation$\sigma$ of the normal perturbation. -
eps: The radius of the balls$B_\epsilon(D_{t-1})$ and$B_\epsilon^{\sigma^2}(D_{t-1})$ . -
nReps: Number of$n$ -point samples in the initial uniform sampling. -
nSteps: Number of steps that the random walk will take. -
k,CoresRequested,save_to_file,results_file: Same as in the main script.
The scripts produces two sets of variables: confs_0, dms_0, bd_times_0, and confs_n, dms_n, bd_times_n. The the initial uniform sample and the random walk are stored in the variables with subindex 0 and n, respectively. The description of dms_* and bd_times_* is the same as in the main script. The difference is confs_*. In this script, each page confs_*(:,:,i) is an
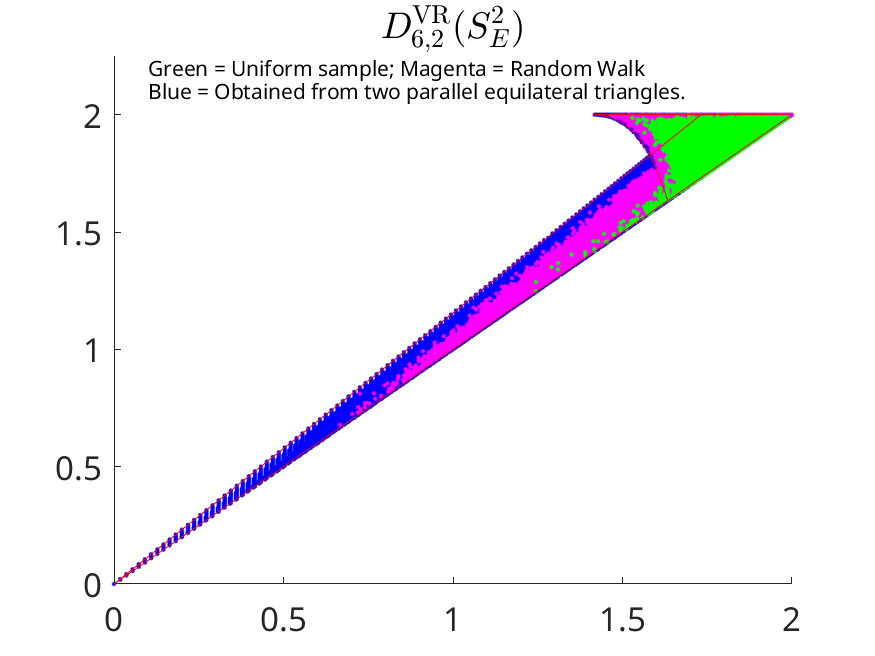 An image similar to the above can be produced by running Persistence_complete_w_rw.m and Plot_S2_conjecture.m in succession with the current parameters.
An image similar to the above can be produced by running Persistence_complete_w_rw.m and Plot_S2_conjecture.m in succession with the current parameters.
The script Persistence_random_walk_plot_progress.m loads a file with a uniform sample of
- Do we want to save the results to a .mat file directly from Persistence_parallel_*.m, or should we gather the results distributed in the checkpoint_temp_file_*.mat files?
- For now, we're saving the results directly to a .mat file. The checkpoints are there for redundancy in case the main program crashes.
- In any case, it would be useful to upload the code that gathers the results from the checkpoints.
- Write a version of Persistence_parallel_Ripser.m that allows for
$n > 2k+2$ . - Set a random seed. This is not straightforward with parallel code because each worker has its own random seed.