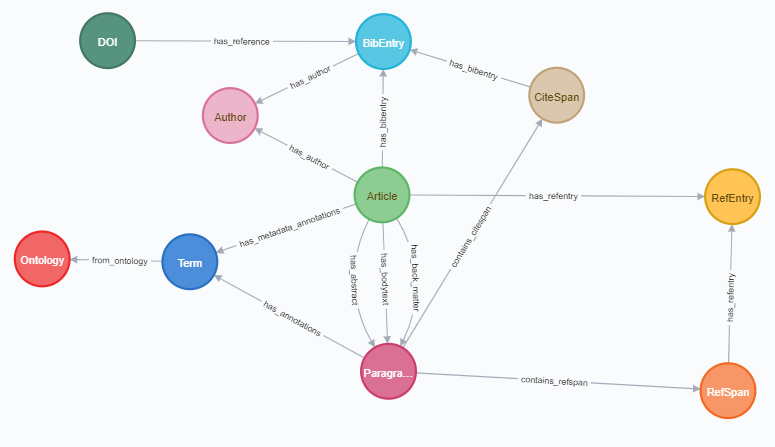
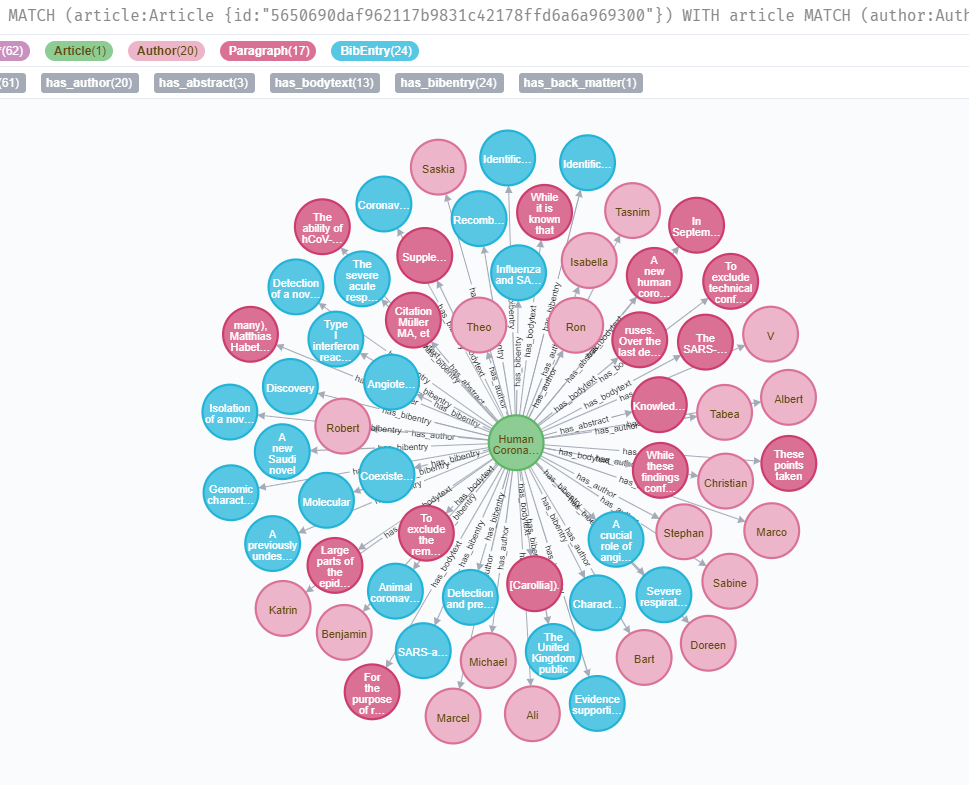
This project takes the annotated dataset from CORD19 project. Original (non-annotated) dataset CORD19 is found in this website.
Our work is part of the Knowledge Graph task for the virtual COVID-19 Biohackathon.
The dataset JSON schema is the following. Our current model is:
Each node has the following properties and labels (node_properties.csv):
| nodeType | nodeLabels | propertyName | propertyTypes | mandatory |
|---|---|---|---|---|
:Author |
[Author] | middle | [StringArray] | true |
:Author |
[Author] | last | [String] | true |
:Author |
[Author] | suffix | [String] | true |
:Author |
[Author] | first | [String] | true |
:Author |
[Author] | [String] | false | |
:Term |
[Term] | id | [String] | true |
:Term |
[Term] | name | [String] | true |
:Ontology |
[Ontology] | name | [String] | true |
:Paragraph |
[Paragraph] | id | [String] | true |
:Paragraph |
[Paragraph] | position | [Long] | true |
:Paragraph |
[Paragraph] | text | [String] | true |
:Paragraph |
[Paragraph] | section | [String] | true |
:CiteSpan |
[CiteSpan] | text | [String] | true |
:CiteSpan |
[CiteSpan] | start | [Long] | true |
:CiteSpan |
[CiteSpan] | end | [Long] | true |
:CiteSpan |
[CiteSpan] | ref_id | [String] | false |
:RefSpan |
[RefSpan] | text | [String] | true |
:RefSpan |
[RefSpan] | start | [Long] | true |
:RefSpan |
[RefSpan] | end | [Long] | true |
:RefSpan |
[RefSpan] | ref_id | [String] | false |
:BibEntry |
[BibEntry] | id | [String] | true |
:BibEntry |
[BibEntry] | title | [String] | true |
:BibEntry |
[BibEntry] | volume | [String] | true |
:BibEntry |
[BibEntry] | venue | [String] | true |
:BibEntry |
[BibEntry] | pages | [String] | true |
:BibEntry |
[BibEntry] | issn | [String] | true |
:BibEntry |
[BibEntry] | year | [Long] | false |
:RefEntry |
[RefEntry] | id | [String] | true |
:RefEntry |
[RefEntry] | text | [String] | true |
:RefEntry |
[RefEntry] | type | [String] | true |
:DOI |
[DOI] | id | [String] | true |
:Article |
[Article] | id | [String] | true |
:Article |
[Article] | title | [String] | true |
:Article |
[Article] | license | [String] | true |
Each edge has the following properties (edges_properties.csv):
| relType | propertyName | propertyTypes | mandatory |
|---|---|---|---|
:has_author |
null | null | false |
:has_metadata_annotations |
hit_sentence_start | [Long] | true |
:has_metadata_annotations |
hit_sentence_end | [Long] | true |
:has_metadata_annotations |
hit_sentences | [Long] | true |
:from_ontology |
null | null | false |
:has_annotations |
hit_sentence_start | [Long] | true |
:has_annotations |
hit_sentence_end | [Long] | true |
:has_annotations |
hit_sentences | [Long] | true |
:has_abstract |
null | null | false |
:has_bodytext |
null | null | false |
:contains_citespan |
null | null | false |
:contains_refspan |
null | null | false |
:has_bibentry |
null | null | false |
:has_refentry |
null | null | false |
:has_back_matter |
null | null | false |
:has_reference |
null | null | false |
CALL db.index.fulltext.queryNodes('termIndex', '<Input>~')
YIELD node, score
RETURN node ORDER BY score DESC SKIP <PAGE> LIMIT <PAGE_SIZE>;Match (t:Term {id:"<TermID>"})<-[*..2]-(a:Article)
WITH a
MATCH (a)-[:has_abstract]->(abstract:Paragraph)
RETURN a, abstract SKIP <PAGE> LIMIT <PAGE_SIZE>;Match (t:Term)<-[r:has_annotations]-(p:Paragraph)<-[*..2]-(a:Article {id:"<ArticleID>"})
RETURN t SKIP <PAGE> LIMIT <PAGE_SIZE>;Match (t:Term {id:"<TermID>"})<-[r:has_annotations]-(p:Paragraph)<-[*..2]-(a:Article {id:"<ArticleID>"})
RETURN t, p, COLLECT(r) as spans ORDER BY p.position ASC SKIP <PAGE> LIMIT <PAGE_SIZE>;MATCH (article:Article {id:"<ArticleID>"})
WITH article
MATCH (author:Author)<-[:has_author]-(article)
WITH article, COLLECT(author) as authors
MATCH (abstract:Paragraph)<-[:has_abstract]-(article)
WITH article, authors, COLLECT(abstract) as abstracts
MATCH (bodytext:Paragraph)<-[:has_bodytext]-(article)
WITH article, authors, abstracts, bodytext
ORDER BY bodytext.position ASC
WITH article, authors, abstracts, COLLECT(bodytext) as bodytexts
MATCH (backmatter:Paragraph)<-[:has_back_matter]-(article)
WITH article, authors, abstracts, bodytexts, COLLECT(backmatter) as backmatters
MATCH (bibEntry:BibEntry)<-[:has_bibentry]-(article)
RETURN article, authors, abstracts, bodytexts, backmatters, COLLECT(bibEntry) as bibEntriesMatch (t:Term)<-[*..2]-(a:Article)
WHERE (t.id IN ["<TermID1>", "<TermID2>"])
WITH a, COUNT(a) as articleCount
MATCH (a)-[:has_metadata_annotations]->(annotation:Term)-[:from_ontology]->(o:Ontology)
RETURN sum(articleCount) as totalArticles, o as ontology, collect(distinct(annotation)) as annotationsThe latter query the article count is divided by the ontology: X articles from Y ontology with Z annotations.
If the ontology is not necessary, you can omit o as ontology, so that the output is: X total articles appearing the Z annotations found.
Match (t:Term)<-[*..2]-(a:Article)
WHERE (t.id IN ["<TermID1>", "<TermID2>"])
WITH a, COUNT(a) as articleCount
MATCH (a)-[:has_metadata_annotations]->(annotation:Term)
RETURN sum(articleCount) as totalArticles, collect(distinct(annotation)) as annotations